Mechanisms of chiral discrimination by topoisomerase IV - PubMed (original) (raw)
Mechanisms of chiral discrimination by topoisomerase IV
K C Neuman et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Topoisomerase IV (Topo IV), an essential ATP-dependent bacterial type II topoisomerase, transports one segment of DNA through a transient double-strand break in a second segment of DNA. In vivo, Topo IV unlinks catenated chromosomes before cell division and relaxes positive supercoils generated during DNA replication. In vitro, Topo IV relaxes positive supercoils at least 20-fold faster than negative supercoils. The mechanisms underlying this chiral discrimination by Topo IV and other type II topoisomerases remain speculative. We used magnetic tweezers to measure the relaxation rates of single and multiple DNA crossings by Topo IV. These measurements allowed us to determine unambiguously the relative importance of DNA crossing geometry and enzymatic processivity in chiral discrimination by Topo IV. Our results indicate that Topo IV binds and passes DNA strands juxtaposed in a nearly perpendicular orientation and that relaxation of negative supercoiled DNA is perfectly distributive. Together, these results suggest that chiral discrimination arises primarily from dramatic differences in the processivity of relaxing positive and negative supercoiled DNA: Topo IV is highly processive on positively supercoiled DNA, whereas it is perfectly distributive on negatively supercoiled DNA. These results provide fresh insight into topoisomerase mechanisms and lead to a model that reconciles contradictory aspects of previous findings while providing a framework to interpret future results.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
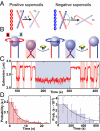
Fig. 1.
Single DNA crossing assay. (A) Crossing geometry for left- and right-handed supercoils. The crossing angle is the clockwise angle between the top and bottom DNA segments. Plectonemes in (+)sc DNA (red) impose an acute angle on juxtaposed DNA segments, whereas plectonemes in (−)sc DNA (blue) impose an obtuse crossing angle on juxtaposed DNA segments. The chirality-sensing model is illustrated with reference to the single crossings. For a preferred crossing angle, α0 < 90°, a small thermal angular fluctuation (δL = α0 − α) is required to relax a left-handed crossing, whereas a larger, hence less probable, fluctuation (δR = 180°−α0 − α), is required to relax a right-handed crossing. (B) Diagram of the single-crossing experimental geometry (not to scale). A paramagnetic bead (blue) is tethered to the surface of a capillary by two DNA molecules (red and blue). Magnets above the capillary apply a constant vertical force on the bead. Rotating the magnets by one clockwise (negative) turn imposes a left-handed crossing and reduces the height of the bead (red shading). Topo IV relaxes the crossing, returning the bead to its original height. The process is repeated by rotating the magnets in the counterclockwise (positive) direction, imposing a right-handed crossing (blue shading), which is subsequently relaxed. (C) Single DNA crossing relaxation measurements. The extension of a doubly tethered bead (red line) is displayed as a function of time for 3.5-μm DNA tethers separated by 0.73 μm at a force of 0.87 pN. Left-handed DNA crossings were relaxed rapidly (red shading), whereas the right-handed crossing took longer to unlink (blue shading). (D) Normalized distribution of relaxation times for left-handed crossings (Left, red bars) and right-handed crossings (Right, blue bars) were fit with single exponentials, P(t) = τ−1exp(−t/τ). The characteristic relaxation times were τL = 2.7 ± 0.6 s (χ2ν = 0.6), and τR = 270 ± 130 s (χ2ν = 0.2), and their ratio was τL/τR = 0.010 ± 0.005.
Fig. S3
displays an example of relaxation times obtained for a different tether geometry and force.
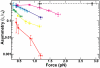
Fig. 2.
Single DNA crossing relaxation rate asymmetry. Single DNA crossing relaxation rate asymmetry (τL/τR) as a function of force for Topo IV (solid colored lines and filled circles) and S. cerevisiae Topo II (dashed black line and open squares) was plotted on a semilogarithmic axis. Error bars represent the propagation of statistical uncertainties. Lines between points are provided to guide the eye. Each tether is characterized by the ratio of the tether separation to the length of the tethers (2_e_/L_0). The 2_e/_L_0 values are: 0.21 (red), 0.27 (yellow), 0.34 (green), 0.44 (cyan), 0.50 (blue), 0.56 (magenta), and 0.25 (black dashed line).
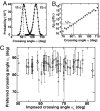
Fig. 3.
Preferred crossing angle (α0) for Topo IV. (A) Angular probability distribution functions, P(α), for a single left- (filled circles) and right-handed (open circles) DNA crossing from MC simulations (12) for the tether geometry and force given in Fig. 1. The angular distribution for the right-handed crossing _P_R(α) was derived from the distribution for the left-handed crossing _P_L(α) through the relation _P_R(α) = _P_L(180° − α). The imposed DNA crossing angle, αL [corresponding to the peak of _P_L(α)] ≈ 59°. (B) Ratio of right-handed to left-handed probability from A plotted on a semilogarithmic axis. The preferred crossing angle, α0, is obtained from the relation τL/τR = _P_R(α0)/_P_L(α0), as illustrated by the gray arrow on the graph. The measured relaxation rate ratio for this tether geometry and force (Fig. 1) was τL/τR = 0.010 ± 0.005, which corresponds to a preferred crossing angle α0 = 82° ± 1.2°.
Fig. S3 C and D
provides an example of the determination of the preferred crossing angle for a larger crossing angle. (C) Preferred crossing angles (α0) for two different Topo IV preparations (open and filled circles) plotted versus the imposed left-handed crossing angle, αL. The combined average preferred crossing angle was α0 = 85.5° ± 0.4° (mean ± SEM). Error bars are a combination of measurement and statistical uncertainties added in quadrature.
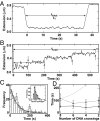
Fig. 4.
Supercoil relaxation by Topo IV. (A) Relaxation of positive supercoils. Extension data were recorded at 25 Hz and median-filtered over 45 points. The extension of the DNA molecule (solid line) decreased rapidly as eight positive supercoils were introduced. All eight supercoils were relaxed in a single burst of activity ≈30 s after they were introduced. The initial time (_t_1) and the total relaxation time (t_total) are indicated. (B) Relaxation of negative supercoils. The extension of the DNA molecule (solid line) rapidly decreased as eight negative supercoils were introduced. Subsequently, individual strand passage events relaxed pairs of supercoils (vertical arrows). (C) Normalized distribution of waiting times for the relaxation of negative supercoils was binned and fit to a single exponential, P(t) = τ−1exp(−_t/τ), (line) (χ2ν = 1.25). (Inset) Distribution of strand passages per binding event on (−)sc DNA. (D) Average time between relaxation events as a function of the number of DNA crossings. The same data from C (open circles and solid line) are compared with data at a 2-fold higher protein concentration (≈150 pM, filled circles and dashed line). For comparison, a model in which the time between relaxation events is inversely proportional to the number of DNA crossings is shown (thin dashed line).
Similar articles
- Chiral discrimination and writhe-dependent relaxation mechanism of human topoisomerase IIα.
Seol Y, Gentry AC, Osheroff N, Neuman KC. Seol Y, et al. J Biol Chem. 2013 May 10;288(19):13695-703. doi: 10.1074/jbc.M112.444745. Epub 2013 Mar 18. J Biol Chem. 2013. PMID: 23508957 Free PMC article. - Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases.
Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Stone MD, et al. Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8654-9. doi: 10.1073/pnas.1133178100. Epub 2003 Jul 11. Proc Natl Acad Sci U S A. 2003. PMID: 12857958 Free PMC article. - Alteration of Escherichia coli topoisomerase IV conformation upon enzyme binding to positively supercoiled DNA.
Crisona NJ, Cozzarelli NR. Crisona NJ, et al. J Biol Chem. 2006 Jul 14;281(28):18927-32. doi: 10.1074/jbc.M603068200. Epub 2006 May 9. J Biol Chem. 2006. PMID: 16684778 - What makes a type IIA topoisomerase a gyrase or a Topo IV?
Hirsch J, Klostermeier D. Hirsch J, et al. Nucleic Acids Res. 2021 Jun 21;49(11):6027-6042. doi: 10.1093/nar/gkab270. Nucleic Acids Res. 2021. PMID: 33905522 Free PMC article. Review. - The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage.
Gubaev A, Klostermeier D. Gubaev A, et al. DNA Repair (Amst). 2014 Apr;16:23-34. doi: 10.1016/j.dnarep.2014.01.011. Epub 2014 Feb 22. DNA Repair (Amst). 2014. PMID: 24674625 Review.
Cited by
- Topo IV is the topoisomerase that knots and unknots sister duplexes during DNA replication.
López V, Martínez-Robles ML, Hernández P, Krimer DB, Schvartzman JB. López V, et al. Nucleic Acids Res. 2012 Apr;40(8):3563-73. doi: 10.1093/nar/gkr1237. Epub 2011 Dec 19. Nucleic Acids Res. 2012. PMID: 22187153 Free PMC article. - DNA self-assembly: from chirality to evolution.
Timsit Y. Timsit Y. Int J Mol Sci. 2013 Apr 15;14(4):8252-70. doi: 10.3390/ijms14048252. Int J Mol Sci. 2013. PMID: 23591841 Free PMC article. Review. - Quantitative guidelines for force calibration through spectral analysis of magnetic tweezers data.
te Velthuis AJ, Kerssemakers JW, Lipfert J, Dekker NH. te Velthuis AJ, et al. Biophys J. 2010 Aug 9;99(4):1292-302. doi: 10.1016/j.bpj.2010.06.008. Biophys J. 2010. PMID: 20713015 Free PMC article. - Grid diagrams as tools to investigate knot spaces and topoisomerase-mediated simplification of DNA topology.
Barbensi A, Celoria D, Harrington HA, Stasiak A, Buck D. Barbensi A, et al. Sci Adv. 2020 Feb 26;6(9):eaay1458. doi: 10.1126/sciadv.aay1458. eCollection 2020 Feb. Sci Adv. 2020. PMID: 32133398 Free PMC article. - The dynamic interplay between DNA topoisomerases and DNA topology.
Seol Y, Neuman KC. Seol Y, et al. Biophys Rev. 2016 Nov;8(Suppl 1):101-111. doi: 10.1007/s12551-016-0240-8. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28510219 Free PMC article. Review.
References
- Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. - PubMed
- Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. - PubMed
- Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. - PubMed
- Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: The story of a simple molecular machine. Q Rev Biophys. 1998;31:107–144. - PubMed
- Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol Microbiol. 2004;52:925–931. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources