Early endosomes and endosomal coatomer are required for autophagy - PubMed (original) (raw)
Early endosomes and endosomal coatomer are required for autophagy
Minoo Razi et al. J Cell Biol. 2009.
Abstract
Autophagy, an intracellular degradative pathway, maintains cell homeostasis under normal and stress conditions. Nascent double-membrane autophagosomes sequester and enclose cytosolic components and organelles, and subsequently fuse with the endosomal pathway allowing content degradation. Autophagy requires fusion of autophagosomes with late endosomes, but it is not known if fusion with early endosomes is essential. We show that fusion of AVs with functional early endosomes is required for autophagy. Inhibition of early endosome function by loss of COPI subunits (beta', beta, or alpha) results in accumulation of autophagosomes, but not an increased autophagic flux. COPI is required for ER-Golgi transport and early endosome maturation. Although loss of COPI results in the fragmentation of the Golgi, this does not induce the formation of autophagosomes. Loss of COPI causes defects in early endosome function, as both transferrin recycling and EGF internalization and degradation are impaired, and this loss of function causes an inhibition of autophagy, an accumulation of p62/SQSTM-1, and ubiquitinated proteins in autophagosomes.
Figures
Figure 1.
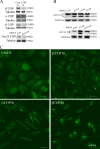
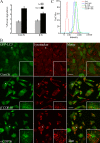
siRNA depletion of COPI increases autophagy. (A) siRNA depletion of β′-, β-, and α-COPI (top) and Sec23A and B (bottom) in 293/GFP-LC3 cells for 48 h. Decreased protein levels of the corresponding subunits is shown by immunoblots. (B) Loss of β′-, α-, and β-COP but not subunits of COPII (Sec 23A and B) increase GFP-LC3-II. In A and B, tubulin was the loading control. (C) GFP-LC3–positive vesicles were detectable in 293/GFP-LC3 cells after siRNA depletion of β′-, α-, and β-COP but only basal levels of GFP-LC3–positive vesicles are observed in control siRNA-depleted cells.
Figure 2.
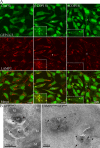
Time course of COPI depletion shows that Golgi disperses before AVs are formed, but Golgi dispersal does not cause AV formation. (A) COPI subunits were depleted in 293/GFP-LC3 cells and analyzed 24, 36, and 48 h after addition of siRNA. Immunoblotting for β′- and α-COP confirms loss of COP subunits. (B) GFP-LC3-I and -II were monitored by immunoblots in parallel lysates using anti-GFP antibodies at the indicated times. (C) After 24 h siRNA treatment, depletion of β′-COP caused morphological changes and a reduction in perinuclear population of GM130. Box indicates enlarged area. (D) Rab1a/b was depleted using siRNAs for 48 h. Lysates were probed with anti-Rab1 to confirm depletion. (E) β′-COP or Rab1a/b were depleted as in D, and analyzed by indirect immunofluorescence using anti-GM130 and GFP fluorescence. In β′-COP panel, the asterisk indicates cells that did not show an accumulation of GFP-LC3–positive AVs.
Figure 3.
AV formation after siRNA depletion of COPI requires Atg5 and Atg7. 293/GFP-LC3 cells were incubated with siRNA for β′-, α-COP, or control siRNA alone or combined with siRNA against Atg5 or Atg7 for 48 h, then (A) lysed and analyzed by immunoblot for Atg5, Atg7, GFP-LC3, and tubulin (loading control), or (B) fixed for immunofluorescence analysis using anti-ERGIC-53 antibodies, or GFP fluorescence. In A, the asterisk indicates nonspecific band; in B and C, asterisk indicates cells that did not show a fragmented ERGIC-53.
Figure 4.
Correlative light-electron microscopy analysis of GFP-LC3 puncta demonstrates they are AVs. (A) Phase, (B) fluorescent, and (C) TEM of the same field of 293/GFP-LC3 cells 48 h after siRNA depletion of α-COP. In B the image was inverted to allow better visualization of the GFP-LC3–positive structures. (D) Higher magnification of cell shown with asterisk in A and C. Top region of cell of interest is enlarged in E to show the two AVs structures indicated by arrows in B. (F and G) Higher magnification of AVs shown by arrows in E.
Figure 5.
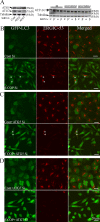
Ub and p62 accumulate in α- and β′-COP–depleted 293/GFP-LC3 cells. (A) 293/GFP-LC3 cells were treated with control or β′-COP siRNA for 48 h, then fixed and labeled with anti-p62 and anti-ubiquitin antibodies for colocalization with GFP-LC3. (B) HEK293 cells treated as in A and labeled with anti-ubiquitin and anti-p62 antibodies. (C) Cryosections from β′-COP–depleted cells were labeled with anti-Ub antibodies followed by 10-nm protein A–gold. AV, autophagosomes.
Figure 6.
AVs in COPI-depleted cells are not degradative. (A) Long-lived protein degradation was assessed 72 h after transfection with control or β′-COP siRNA in 293/GFP-LC3 cells. Cells were either starved of amino acids for 2 h (St) or incubated in fresh growth medium (Fed) for 2 h. The amount of protein degradation is presented as mean ± SEM. (B) siControl-treated cells and β′- and α-COP were depleted for 48 h, then incubated with Lysotracker red (50 nM) for 30 min before fixation. Merged image shows very little colocalization of GFP-LC3 AV-positive structures. Asterisk shows cell enlarged in inset. (C) After β′- and α-COP siRNA depletion, Magic Red-RR2 (MR) was used to detect active cathepsin B in cells which were fed or starved (St) for 2 h. The intensity of MR labeling was measured in live cells by flow cytometry.
Figure 7.
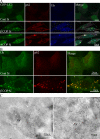
GFP-LC3 AVs are LAMP2 positive. (A) 293/GFP-LC3 cells were incubated with control or β′- or α-COP siRNA for 48 h, fixed and labeled with anti-LAMP2 antibodies, followed by Alexa 555 anti–mouse antibodies. After COPI depletion many GFP-LC3–positive AVs are positive for LAMP2. Asterisk indicates cell enlarged in inset. (B) Cryosections of β′-COP-depleted cells labeled with anti-GFP (left) or anti-GFP and anti-LAMP2 (right) followed by protein A–gold as indicated. AV, autophagosomes, M, mitochondria.
Figure 8.
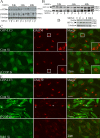
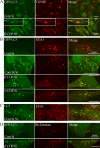
GFP-LC3–positive AVs colocalize TGN46 and early endocytotic markers after COPI depletion. (A) 293/GFP-LC3 cells were incubated with siRNA for β′-COP or control siRNA (Cont) for 48 h, processed for immunofluorescence, and labeled with antibodies for TGN46. In (B) siRNA-treated cells were fixed and labeled, or starved for 2 h (St), and labeled with antibodies to EEA1. (C) Control-starved HEK293 cells were labeled for endogenous LC3 and EEA1 by double labeling after methanol fixation. (D) After siRNA treatment, rhodamine-labeled dextran was internalized for 20 min, followed by a 40-min chase after which the 293/GFP-LC3 cells were fixed and analyzed by confocal microscopy.
Figure 9.
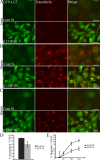
Transferrin and EGF trafficking is inhibited by COPI depletion. 293/GFP-LC3 cells were treated with β′-COP siRNA or control siRNA. After 48 h cells were incubated with Alexa 555 transferrin (A) for 2 min, (B) 30 min, or (C) 30 min followed by a 30-min chase, then washed and fixed and analyzed by confocal microscopy. (D and E) 125I-EGF was added to HeLa cells after 48 h of siRNA depletion of β′-COP, or control siRNA for 10 min. The cells were then washed as described in the Materials and methods, and chased for 30, 60, 120, and 180 min (including 10-min incubation). (D) The amount of 125I-EGF internalized was shown as a percentage of the total 125I-EGF taken up in 10 min. (E) The amount of degraded 125I-EGF was determined as a percentage of the total radioactivity present after TCA precipitation over the total 125I-EGF internalized. The experiment was performed three times in duplicate and the data shown is the mean ± SEM. The significance was determined by Student's t test. *, P ≤ 0.05.
Similar articles
- PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I. McEwan DG, et al. Mol Cell. 2015 Jan 8;57(1):39-54. doi: 10.1016/j.molcel.2014.11.006. Epub 2014 Dec 11. Mol Cell. 2015. PMID: 25498145 - VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD.
Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. Tresse E, et al. Autophagy. 2010 Feb;6(2):217-27. doi: 10.4161/auto.6.2.11014. Epub 2010 Feb 22. Autophagy. 2010. PMID: 20104022 Free PMC article. - In vitro reconstitution of fusion between immature autophagosomes and endosomes.
Morvan J, Köchl R, Watson R, Collinson LM, Jefferies HB, Tooze SA. Morvan J, et al. Autophagy. 2009 Jul;5(5):676-89. doi: 10.4161/auto.5.5.8378. Epub 2009 Jul 6. Autophagy. 2009. PMID: 19337031 - Autophagy and multivesicular bodies: two closely related partners.
Fader CM, Colombo MI. Fader CM, et al. Cell Death Differ. 2009 Jan;16(1):70-8. doi: 10.1038/cdd.2008.168. Epub 2008 Nov 14. Cell Death Differ. 2009. PMID: 19008921 Review. - Biological Roles of Alternative Autophagy.
Shimizu S. Shimizu S. Mol Cells. 2018 Jan 31;41(1):50-54. doi: 10.14348/molcells.2018.2215. Epub 2018 Jan 23. Mol Cells. 2018. PMID: 29370693 Free PMC article. Review.
Cited by
- Maturing reticulocytes internalize plasma membrane in glycophorin A-containing vesicles that fuse with autophagosomes before exocytosis.
Griffiths RE, Kupzig S, Cogan N, Mankelow TJ, Betin VM, Trakarnsanga K, Massey EJ, Lane JD, Parsons SF, Anstee DJ. Griffiths RE, et al. Blood. 2012 Jun 28;119(26):6296-306. doi: 10.1182/blood-2011-09-376475. Epub 2012 Apr 6. Blood. 2012. PMID: 22490681 Free PMC article. - Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome.
Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Tumbarello DA, et al. Nat Cell Biol. 2012 Oct;14(10):1024-35. doi: 10.1038/ncb2589. Epub 2012 Sep 30. Nat Cell Biol. 2012. PMID: 23023224 Free PMC article. - COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis.
Watkin LB, Jessen B, Wiszniewski W, Vece TJ, Jan M, Sha Y, Thamsen M, Santos-Cortez RL, Lee K, Gambin T, Forbes LR, Law CS, Stray-Pedersen A, Cheng MH, Mace EM, Anderson MS, Liu D, Tang LF, Nicholas SK, Nahmod K, Makedonas G, Canter DL, Kwok PY, Hicks J, Jones KD, Penney S, Jhangiani SN, Rosenblum MD, Dell SD, Waterfield MR, Papa FR, Muzny DM, Zaitlen N, Leal SM, Gonzaga-Jauregui C; Baylor-Hopkins Center for Mendelian Genomics; Boerwinkle E, Eissa NT, Gibbs RA, Lupski JR, Orange JS, Shum AK. Watkin LB, et al. Nat Genet. 2015 Jun;47(6):654-60. doi: 10.1038/ng.3279. Epub 2015 Apr 20. Nat Genet. 2015. PMID: 25894502 Free PMC article. - Photothermal exposure of polydopamine-coated branched Au-Ag nanoparticles induces cell cycle arrest, apoptosis, and autophagy in human bladder cancer cells.
Zhao X, Qi T, Kong C, Hao M, Wang Y, Li J, Liu B, Gao Y, Jiang J. Zhao X, et al. Int J Nanomedicine. 2018 Oct 12;13:6413-6428. doi: 10.2147/IJN.S174349. eCollection 2018. Int J Nanomedicine. 2018. PMID: 30410328 Free PMC article. - mTORC1 controls Golgi architecture and vesicle secretion by phosphorylation of SCYL1.
Kaeser-Pebernard S, Vionnet C, Mari M, Sankar DS, Hu Z, Roubaty C, Martínez-Martínez E, Zhao H, Spuch-Calvar M, Petri-Fink A, Rainer G, Steinberg F, Reggiori F, Dengjel J. Kaeser-Pebernard S, et al. Nat Commun. 2022 Aug 10;13(1):4685. doi: 10.1038/s41467-022-32487-7. Nat Commun. 2022. PMID: 35948564 Free PMC article.
References
- Bampton E.T.W., Goemans C.G., Niranjan D., Mizushima N., Tolkovsky A.M. 2005. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes.Autophagy. 1:23–36 - PubMed
- Boyce M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D., Yuan J. 2005. A selective inhibitor of eIF2{alpha} dephosphorylation protects cells from ER stress.Science. 307:935–939 - PubMed
- Chan E.Y.W., Kir S., Tooze S.A. 2007. siRNA screening of the kinome identifies ULK1 as a multi-domain modulator of autophagy.J. Biol. Chem. 282:25464–25474 - PubMed
- Chapuy B., Tikkanen R., Muhlhausen C., Wenzel D., von Figura K., Honing S. 2008. AP-1 and AP-3 mediate sorting of melanosomal and lysosomal membrane proteins into distinct post-Golgi trafficking pathways.Traffic. 9:1157–1172 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous