Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria - PubMed (original) (raw)
Comparative Study
. 2009 Apr 14;6(4):e1000055.
doi: 10.1371/journal.pmed.1000055. Epub 2009 Apr 14.
Hirva Pota, Marie-Solange B Evehe, El-Hadj Bâ, Ghyslain Mombo-Ngoma, Allen L Malisa, Rosalynn Ord, Walter Inojosa, Alexandre Matondo, Diadier A Diallo, Wilfred Mbacham, Ingrid V van den Broek, Todd D Swarthout, Asefaw Getachew, Seyoum Dejene, Martin P Grobusch, Fanta Njie, Samuel Dunyo, Margaret Kweku, Seth Owusu-Agyei, Daniel Chandramohan, Maryline Bonnet, Jean-Paul Guthmann, Sian Clarke, Karen I Barnes, Elizabeth Streat, Stark T Katokele, Petrina Uusiku, Chris O Agboghoroma, Olufunmilayo Y Elegba, Badara Cissé, Ishraga E A-Elbasit, Hayder A Giha, S Patrick Kachur, Caroline Lynch, John B Rwakimari, Pascalina Chanda, Moonga Hawela, Brian Sharp, Inbarani Naidoo, Cally Roper
Affiliations
- PMID: 19365539
- PMCID: PMC2661256
- DOI: 10.1371/journal.pmed.1000055
Comparative Study
Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria
Richard J Pearce et al. PLoS Med. 2009.
Abstract
Background: Although the molecular basis of resistance to a number of common antimalarial drugs is well known, a geographic description of the emergence and dispersal of resistance mutations across Africa has not been attempted. To that end we have characterised the evolutionary origins of antifolate resistance mutations in the dihydropteroate synthase (dhps) gene and mapped their contemporary distribution.
Methods and findings: We used microsatellite polymorphism flanking the dhps gene to determine which resistance alleles shared common ancestry and found five major lineages each of which had a unique geographical distribution. The extent to which allelic lineages were shared among 20 African Plasmodium falciparum populations revealed five major geographical groupings. Resistance lineages were common to all sites within these regions. The most marked differentiation was between east and west African P. falciparum, in which resistance alleles were not only of different ancestry but also carried different resistance mutations.
Conclusions: Resistant dhps has emerged independently in multiple sites in Africa during the past 10-20 years. Our data show the molecular basis of resistance differs between east and west Africa, which is likely to translate into differing antifolate sensitivity. We have also demonstrated that the dispersal patterns of resistance lineages give unique insights into recent parasite migration patterns.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
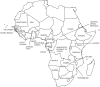
Figure 1. Map of the countries of Africa included in this study.
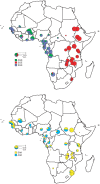
Figure 2. The distribution of the major dhps alleles across sub-Saharan Africa.
Resistant alleles; the upper map shows the relative proportions of the three major resistance alleles, SGK, AGK, and SGE. Wild-type alleles; the lower map shows the ratio of SAK and AAK alleles among wild-type dhps alleles. In both cases the diameter of the pie is proportional to the combined frequencies of the alleles represented in the total population.
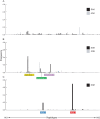
Figure 3. Microsatellite diversity around the wild-type (SAK and AAK) and the resistant (AGK, SGK, and SGE) alleles.
The expected heterozygosity (H e) at flanking loci 0.8 kb, 4.3 kb, and 7.7 kb from the dhps gene was calculated for each geographical site (provided the number of observation ≥10), and box plots show the median, interquartile ranges, and the upper and lower extremes of the distribution of H e values among geographical sites. Where there are statistical outliers, these are indicated by small squares.
Figure 4. Microsatellite polymorphism flanking wild-type and resistant dhps alleles.
In the bar graphs all the microsatellite haplotypes observed have been ranked first according to allele size at locus 0.8 kb and then by allele size at locus 4.3 kb along a common _x_-axis. The association of specific microsatellite haplotypes with different dhps alleles is apparent from the frequencies of each haplotype shown in the individual charts. (A) haplotypes linked to SAK AAK wild-type alleles, (B) haplotypes linked to AGK SGK single-mutant alleles, and (C) haplotypes linked to SGE double-mutant alleles.
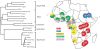
Figure 5. The African distribution of dhps resistance lineages.
The distribution of the five major lineages among the geographic sites is indicated in the map. Resistance alleles whose flanking microsatellite haplotypes did not conform to a defined major lineage are shown in grey. Sharing of resistance allele lineages among the African populations is shown in a cladogram based on pairwise comparison of allele sharing (D PS), which includes all the flanking haplotypes identified. Closely related populations cluster in large geographic regions that supercede national boundaries.
Comment in
- Mapping the spread of malaria drug resistance.
Anderson T. Anderson T. PLoS Med. 2009 Apr 14;6(4):e1000054. doi: 10.1371/journal.pmed.1000054. Epub 2009 Apr 14. PLoS Med. 2009. PMID: 19365538 Free PMC article. - Mapping the origins and spread of antifolate-resistant malaria parasites.
Hyde JE. Hyde JE. Future Microbiol. 2009 Oct;4(8):953-8. doi: 10.2217/fmb.09.66. Future Microbiol. 2009. PMID: 19824787
Similar articles
- Mapping the spread of malaria drug resistance.
Anderson T. Anderson T. PLoS Med. 2009 Apr 14;6(4):e1000054. doi: 10.1371/journal.pmed.1000054. Epub 2009 Apr 14. PLoS Med. 2009. PMID: 19365538 Free PMC article. - Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum.
Naidoo I, Roper C. Naidoo I, et al. Trends Parasitol. 2010 Sep;26(9):447-56. doi: 10.1016/j.pt.2010.05.001. Epub 2010 Jun 17. Trends Parasitol. 2010. PMID: 20728060 Review. - Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in Ghana.
Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, Ghansah A, Asamoa K, Slutsker L, Wilson MD, Barnwell JW, Udhayakumar V, Koram KA. Alam MT, et al. J Infect Dis. 2011 Jan 15;203(2):220-7. doi: 10.1093/infdis/jiq038. J Infect Dis. 2011. PMID: 21288822 Free PMC article. - A multiplex ligase detection reaction-fluorescent microsphere assay for simultaneous detection of single nucleotide polymorphisms associated with Plasmodium falciparum drug resistance.
Carnevale EP, Kouri D, DaRe JT, McNamara DT, Mueller I, Zimmerman PA. Carnevale EP, et al. J Clin Microbiol. 2007 Mar;45(3):752-61. doi: 10.1128/JCM.01683-06. Epub 2006 Nov 22. J Clin Microbiol. 2007. PMID: 17121999 Free PMC article. - Origins and spread of pfdhfr mutant alleles in Plasmodium falciparum.
Mita T. Mita T. Acta Trop. 2010 Jun;114(3):166-70. doi: 10.1016/j.actatropica.2009.07.008. Epub 2009 Jul 14. Acta Trop. 2010. PMID: 19607799 Review.
Cited by
- A snapshot of the prevalence of dihydropteroate synthase-431V mutation and other sulfadoxine-pyrimethamine resistance markers in Plasmodium falciparum isolates in Nigeria.
Adegbola AJ, Ijarotimi OA, Ubom AE, Adesoji BA, Babalola OE, Hocke EF, Hansson H, Mousa A, Bolaji OO, Alifrangis M, Roper C. Adegbola AJ, et al. Malar J. 2023 Mar 1;22(1):71. doi: 10.1186/s12936-023-04487-5. Malar J. 2023. PMID: 36859238 Free PMC article. - Distinctive origin and spread route of pyrimethamine-resistant Plasmodium falciparum in southern China.
Zhang Y, Yan H, Wei G, Han S, Huang Y, Zhang Q, Pan W. Zhang Y, et al. Antimicrob Agents Chemother. 2014;58(1):237-46. doi: 10.1128/AAC.00972-13. Epub 2013 Oct 21. Antimicrob Agents Chemother. 2014. PMID: 24145550 Free PMC article. - Integrating rapid risk mapping and mobile phone call record data for strategic malaria elimination planning.
Tatem AJ, Huang Z, Narib C, Kumar U, Kandula D, Pindolia DK, Smith DL, Cohen JM, Graupe B, Uusiku P, Lourenço C. Tatem AJ, et al. Malar J. 2014 Feb 10;13:52. doi: 10.1186/1475-2875-13-52. Malar J. 2014. PMID: 24512144 Free PMC article. - Whole Genome Sequencing of Field Isolates Reveals Extensive Genetic Diversity in Plasmodium vivax from Colombia.
Winter DJ, Pacheco MA, Vallejo AF, Schwartz RS, Arevalo-Herrera M, Herrera S, Cartwright RA, Escalante AA. Winter DJ, et al. PLoS Negl Trop Dis. 2015 Dec 28;9(12):e0004252. doi: 10.1371/journal.pntd.0004252. eCollection 2015 Dec. PLoS Negl Trop Dis. 2015. PMID: 26709695 Free PMC article. - Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum in Nigeria and the regional emergence of dhps 431V.
Oguike MC, Falade CO, Shu E, Enato IG, Watila I, Baba ES, Bruce J, Webster J, Hamade P, Meek S, Chandramohan D, Sutherland CJ, Warhurst D, Roper C. Oguike MC, et al. Int J Parasitol Drugs Drug Resist. 2016 Dec;6(3):220-229. doi: 10.1016/j.ijpddr.2016.08.004. Epub 2016 Sep 29. Int J Parasitol Drugs Drug Resist. 2016. PMID: 27821281 Free PMC article.
References
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, et al. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. - PubMed
- Su X, Kirkman LA, Fujioka H, Wellems TE. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. - PubMed
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. - PubMed
- Payne D. The History and Development of WHO Standard in vivo and in vitro Test Systems for the Sensitivity of Plasmodium falciparum and other Human Plasmodia to Antimalarial Drugs. London: University of London (London School of Hygiene and Tropical Medicine); 1989.
- Charmot G, Amat-Roze JM, Rodhain F, Le Bras J, Coulaud JP. [Geographic approach to the epidemiology of chloroquine-resistance of Plasmodium falciparum in tropical Africa]. Ann Soc Belg Med Trop. 1991;71:187–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous