Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1 - PubMed (original) (raw)
Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1
Cora L Woodward et al. J Virol. 2009 Jul.
Abstract
The ability to traverse an intact nuclear envelope and productively infect nondividing cells is a salient feature of human immunodeficiency virus type 1 (HIV-1) and other lentiviruses, but the viral factors and mechanism of nuclear entry have not been defined. HIV-1 integrase (IN) is implicated to play a role in the nuclear import of the virus, but the cellular pathway for IN trafficking and the role of IN in mediating the nuclear import of viral particles are unknown. Using a semipermeabilized cell assay, we observed that the nuclear import of IN was not the result of passive diffusion but occurred independently of cytosolic factors, metabolic energy, and the classical receptor-mediated, Ran-dependent import pathways. To determine if IN enters the nucleus by interacting with the nucleopore complex (NPC), we found that IN bound directly with the FxFG-rich C-terminal domain of nucleoporin 153 (NUP153C). When added in excess to the import assay, NUP153C inhibited the nuclear import of IN. Known binding partners of NUP153C competed with IN for binding with NUP153 and also inhibited the nuclear import of IN. In cultured cells, overexpression of NUP153C reduced the infectivity of an HIV-derived vector by interfering with the nuclear translocation of the viral cDNA. These results support a functional role for the IN-NUP153 interaction in HIV-1 replication and suggest that HIV-1 subviral particles gain access to the nucleus by interacting directly with the NPC via the binding of particle-associated IN to NUP153C.
Figures
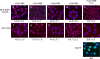
FIG. 1.
The nuclear import of HIV-1 IN is independent of cytosolic factors and metabolic energy. Nuclear import requirements of NLS-BSA-TRITC (a to e) and FLAG-tagged IN (f to j) were characterized using the semipermeabilized cell assay in the presence (+) or absence (−) of cytosolic extract (Cyt) and metabolic energy (ME) at 30°C (a to d and f to i) or at 4°C (e, j, and k). Quantitation for the level of nuclear import observed under each reaction condition was based on ≥50 nuclei. Complete reactions for NLS-BSA-TRITC (a) and HIV-1 IN (f) were set to 100%, and the levels of nuclear import observed (b to e and g to j) were calculated as percentages of the corresponding complete reactions in NLS-BSA-TRITC (a) and IN (f), respectively. Values indicated below the images are the means ± SEM for three experiments and represent the total fluorescence emanating from the import protein localized to the nucleus. NLS-BSA-TRITC was visualized directly by fluorescence microscopy. FLAG-tagged HIV-1 IN was visualized by indirect immunofluorescence using a monoclonal antibody directed at the FLAG epitope, followed by a fluorescently labeled secondary antibody. For uniformity, the fluorescence signal of both HIV-1 IN and NLS-BSA-TRITC was pseudocolored as red, while the blue fluorescence represents the DAPI-counterstained nuclei. Purified recombinant EGFP (27 kDa) was used as a control protein for passive diffusion under import conditions that restricted active transport, and its fluorescence signal was not pseudocolored. ND, not determined.
FIG. 2.
Inhibitors of classical nuclear import pathways do not affect HIV-1 IN nuclear import. (A) Differential effects of the nonhydrolyzable GTP analog GMP-PNP on the nuclear import of NLS-BSA-TRITC and HIV-1 IN. GMP-PNP was included in the import reaction at a final concentration of 2.5 mM. Quantitation of nuclear import observed under each reaction condition was done as described in the legend to Fig. 1 and expressed as percentages of the corresponding reactions without GMP-PNP, as shown in Fig. 1a (NLS-BSA-TRITC) and Fig 1f and i (HIV-1 IN). (B) Differential effects of excess SV40 NLS peptide. Excess purified peptide corresponding to the NLS of the SV40 large T antigen was included in the nuclear import reactions at the indicated concentrations. Quantitation of nuclear import observed under each reaction condition was done as described in the legend to Fig. 1 and expressed as percentages of the corresponding reactions without the peptide competitor, as shown in panels a (NLS-BSA-TRITC) and e (HIV-1 IN). Symbols have the same connotations as described in the legend to Fig. 1. Values are the means ± SEM for three experiments.
FIG. 3.
WGA inhibits the nuclear import of NLS-BSA-TRITC and HIV-1 IN. Nuclear import reactions for NLS-BSA-TRITC and HIV-1 IN were carried out as described in the legend to Fig. 1, except that the cells were pretreated with WGA for 15 min at 30°C. Assay conditions are described along the top of the images, and the quantitation method and symbols are used as described in the legend to Fig. 1. Values are expressed as mean percentages ± SEM of the corresponding reaction carried out in the absence of WGA, as shown in Fig. 1a (NLS-BSA-TRITC) and Fig. 1i (HIV-1 IN). The fluorescence signal for EGFP (c) was not pseudocolored as red.
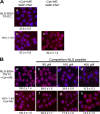
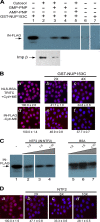
FIG. 4.
HIV-1 IN interacts with NUP153C. (A) HIV-1 IN binds to NUP153C. GST-NUP153C (60 pmol) was loaded onto glutathione-agarose beads and incubated with FLAG-tagged HIV-1 IN (33 pmol; lanes 1 to 5) or with FLAG-tagged FIV IN (33 pmol; lane 7) in binding buffer. Lane 6 shows a control reaction in which NUP153C was omitted. Binding experiments were carried out in the absence (lanes 1, 4, 6, and 7) or presence (lanes 2, 3, and 5) of the HeLa cytosolic extract and with (lanes 3 and 4) or without (lanes 1, 2, 5, 6, and 7) GMP-PNP. AMP-PNP is included as a specificity control for the stimulatory effect of GMP-PNP on the binding of HIV-1 IN to GST-NUP153C in the presence of cytosol (lane 5). Imp β contained in the HeLa cytosolic extract that was bound to NUP153C in the absence (bottom, lane 2) or presence (bottom, lane 3) of GMP-PNP was probed with a monoclonal anti-imp β antibody (Sigma). Arrows indicate the position corresponding to FLAG-tagged HIV-1 IN (top, lanes 1 to 6), FLAG-tagged FIV IN (top, lane 7), and imp β (bottom, lanes 2 and 3). (B) NUP153C inhibits the nuclear import of NLS-BSA-TRITC and HIV-1 IN. Nuclear import reactions for NLS-BSA-TRITC and HIV-1 were carried out under the indicated reaction conditions and as described in the legend to Fig. 1, except that GST-NUP153C was added at a molar ratio of 2:1 (panels b and e) or 4:1 (panels c and f) to the import protein. Quantitation of nuclear import for ≥50 nuclei from each reaction is indicated below the images and reported as the percentage of the corresponding reaction carried out in the absence of excess GST-NUP153C, as shown in panels a (NLS-BSA-TRITC) and d (IN-FLAG). (C) HIV-1 IN binding to NUP153C is inhibited by NTF2. GST pulldown reactions were carried out in the absence of the cytosolic extract (lane 1) and as described above for panel A, except that purified NTF2 was included as a competitive inhibitor for IN binding to NUP153C (lanes 2 to 4). Purified BSA was used as a control (lanes 5 to 7). Ratios indicate the molar excess of competitor protein to IN. (D) NTF2 inhibits the nuclear import of HIV-1 IN. The nuclear import reaction for HIV-1 IN was carried out in the absence of cytosol and metabolic energy, as described in the legend to Fig. 1, except that NTF2 was added as a competitor (panels b to d) to the import reaction at the indicated excesses. Quantitation of nuclear import for ≥50 nuclei from each reaction is indicated below the images and reported as the percentage of the corresponding reaction carried out in the absence of excess NTF2, as shown in panel a. Values are the means ± SEM of three experiments, and symbols have the same connotations as described in the legend to Fig. 1.
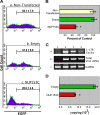
FIG. 5.
Infectivity of a lentiviral-based vector is inhibited by overexpressing NUP153C. (A) The level of EGFP transgene expression from an HIV-1-based vector is reduced in the presence of NUP153C. Nontransfected 293T cells (panel a) and cells transfected with an empty plasmid (panel b) or with a plasmid encoding NUP153C (panel c) were challenged with a VSV-G-pseudotyped HIV-1-based vector encoding EGFP at 8 h posttransfection. Twenty-four hours postinfection, the cells were collected and analyzed by flow cytometry for EGFP expression. Histograms are representative of results under the indicated experimental conditions. The number of EGFP-positive cells under each condition is expressed as the mean ± SEM for at least three experiments. (B) Graphical representation of EGFP expression normalized to the nontransfected control. An asterisk indicates a significant difference from the empty vector control (P = 0.0002). (C) NUP153C expression inhibits the nuclear import of viral cDNA. DNA was extracted from the same cell populations that were analyzed by flow cytometry in panel A and was subjected to PCR using primers to amplify the 1-LTR circle form of viral DNA (top), total viral DNA (middle), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (bottom) from genomic DNA. Lane 1 shows nontransduced 293T cells. Lane 2 shows 293T cells transduced with the VSV-G-pseudotyped HIV-1-based vector encoding EGFP. Lane 3 shows 293T cells transfected with an empty vector 8 h before transduction with the HIV-1 vector. Lane 4 shows 293T cells transfected with NUP153C 8 h prior to transduction with the HIV-1 vector. (D) NUP153C expression inhibits viral cDNA integration. _Alu_-PCR was used to quantify the copy number of provirus integrated in cells transfected with either an empty vector (green bar; n = 3) or the NUP153C-expressing vector (red bar; n = 9). The values are expressed as the copy number of provirus/ng genomic DNA. An asterisk indicates a significant difference from the empty vector control (P = 0.0002).
Similar articles
- The nuclear pore complex: a new dynamic in HIV-1 replication.
Woodward CL, Chow SA. Woodward CL, et al. Nucleus. 2010 Jan-Feb;1(1):18-22. doi: 10.4161/nucl.1.1.10571. Nucleus. 2010. PMID: 21327100 Free PMC article. - Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells.
Buffone C, Martinez-Lopez A, Fricke T, Opp S, Severgnini M, Cifola I, Petiti L, Frabetti S, Skorupka K, Zadrozny KK, Ganser-Pornillos BK, Pornillos O, Di Nunzio F, Diaz-Griffero F. Buffone C, et al. J Virol. 2018 Sep 12;92(19):e00648-18. doi: 10.1128/JVI.00648-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997211 Free PMC article. - Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity.
Matreyek KA, Yücel SS, Li X, Engelman A. Matreyek KA, et al. PLoS Pathog. 2013;9(10):e1003693. doi: 10.1371/journal.ppat.1003693. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24130490 Free PMC article. - A hard way to the nucleus.
Bukrinsky M. Bukrinsky M. Mol Med. 2004 Jan-Jun;10(1-6):1-5. Mol Med. 2004. PMID: 15502876 Free PMC article. Review. - Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV?
Piller SC, Caly L, Jans DA. Piller SC, et al. Curr Drug Targets. 2003 Jul;4(5):409-29. doi: 10.2174/1389450033490984. Curr Drug Targets. 2003. PMID: 12816349 Review.
Cited by
- Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication.
Di Nunzio F, Fricke T, Miccio A, Valle-Casuso JC, Perez P, Souque P, Rizzi E, Severgnini M, Mavilio F, Charneau P, Diaz-Griffero F. Di Nunzio F, et al. Virology. 2013 May 25;440(1):8-18. doi: 10.1016/j.virol.2013.02.008. Epub 2013 Mar 21. Virology. 2013. PMID: 23523133 Free PMC article. - Cellular cofactors of lentiviral integrase: from target validation to drug discovery.
Taltynov O, Desimmie BA, Demeulemeester J, Christ F, Debyser Z. Taltynov O, et al. Mol Biol Int. 2012;2012:863405. doi: 10.1155/2012/863405. Epub 2012 Aug 7. Mol Biol Int. 2012. PMID: 22928108 Free PMC article. - Toward Structurally Novel and Metabolically Stable HIV-1 Capsid-Targeting Small Molecules.
Vernekar SKV, Sahani RL, Casey MC, Kankanala J, Wang L, Kirby KA, Du H, Zhang H, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Vernekar SKV, et al. Viruses. 2020 Apr 16;12(4):452. doi: 10.3390/v12040452. Viruses. 2020. PMID: 32316297 Free PMC article. - HIV-1 remodels the nuclear pore complex.
Monette A, Panté N, Mouland AJ. Monette A, et al. J Cell Biol. 2011 May 16;193(4):619-31. doi: 10.1083/jcb.201008064. J Cell Biol. 2011. PMID: 21576391 Free PMC article. - N-terminally truncated POM121C inhibits HIV-1 replication.
Saito H, Takeuchi H, Masuda T, Noda T, Yamaoka S. Saito H, et al. PLoS One. 2017 Sep 5;12(9):e0182434. doi: 10.1371/journal.pone.0182434. eCollection 2017. PLoS One. 2017. PMID: 28873410 Free PMC article.
References
- Adam, S. A., R. Sterne-Marr, and L. Gerace. 1992. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 21997-110. - PubMed
- Akhtar, A., and S. M. Gasser. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8507-517. - PubMed
- Appa, R. S., C. G. Shin, P. Lee, and S. A. Chow. 2001. Role of the nonspecific DNA-binding region and alpha helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 27645848-45855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA068859/CA/NCI NIH HHS/United States
- AI28697/AI/NIAID NIH HHS/United States
- CA16042/CA/NCI NIH HHS/United States
- CA68859/CA/NCI NIH HHS/United States
- P30 CA016042/CA/NCI NIH HHS/United States
- P30 AI028697/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous