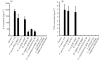Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis - PubMed (original) (raw)
Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis
Toshifumi Ohkusa et al. J Med Microbiol. 2009 May.
Abstract
Interleukin 2 (IL-2)- and IL-10-knockout mice develop spontaneous colitis under conventional but not germ-free conditions, suggesting that commensal bacteria play an important role in the pathogenesis of colitis. However, interactions between commensal bacteria and colonic epithelial cells have not been fully investigated. We therefore assessed the ability of various commensal bacteria and probiotics to adhere to and invade colonic epithelial cells. Effects of the bacteria on production of proinflammatory cytokines were also measured. Commensal bacteria, including mucosal organisms isolated from ulcerative colitis (UC) patients, such as Fusobacterium varium, reported as a possible pathogen in UC, Bacteroides vulgatus, Escherichia coli and Clostridium clostridioforme, as well as their type strains and probiotics, were assessed for their ability to adhere to and invade colonic epithelial cells using two cell lines, SW-480 and HT-29. Our experiments employed co-incubation, a combination of scanning and transmission electron microscopy and recovery of bacteria from infected-cell lysates. F. varium and several other commensal bacteria, but not probiotics, adhered to colonic epithelial cells and invaded their cytoplasm. ELISA and real-time PCR revealed that the host cells, particularly those invaded by F. varium, showed significant increases in IL-8 and TNF-alpha concentrations in supernatants, with elevation of IL-8, TNF-alpha, MCP-1 and IL-6 mRNAs. Furthermore, IL-8 and TNF-alpha expression and nuclear phosphorylated NF-kappaB p65 expression could be immunohistochemically confirmed in inflamed epithelium with cryptitis or crypt abscess in UC patients. Certain commensal bacteria can invade colonic epithelial cells, activating early intracellular signalling systems to trigger host inflammatory reactions.
Figures
Fig. 1.
Adherence and invasion of commensal bacteria and probiotics in SW-480 cells (SEM and TEM). (a) The scanning electron micrographs show attachment of bacteria, with the exception of L. delbrueckii subsp. bulgaricus LB-021001, to SW-480 cells. F. varium 113 was more adhesive than the other bacterial strains tested. Bar, 1 μm. (b) Commensal bacteria, F. varium 113 and B. vulgatus 90, isolated from patients with UC, and E. coli JCM 1649T adhere to SW-480 cells and invade their cytoplasm (arrows), while the probiotic L. delbrueckii subsp. bulgaricus does not enter the cells (TEM photograph; bar, 1 μm).
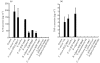
Fig. 2.
Production of cytokines upon infection with commensal bacteria and probiotics in SW-480 cells. (a) IL-8. In the invasion assay, F. varium 113 stimulated the greatest IL-8 production, followed by F. varium ATCC 8501T, E. coli JCM 1649T, B. vulgatus JCM 5826T, B. vulgatus 90 and C. clostridioforme 94. Since the IL-8 levels at the 95 % confidence interval for these bacteria were above those of the controls (<16 pg ml−1), these bacteria significantly stimulated IL-8 production by SW-480. The IL-8 levels with F. varium 113, ATCC 8501T and E. coli JCM 1649T were significantly higher than those with B. vulgatus 90 and C. clostridioforme 94 (P <0.01). (b) TNF-α. In the invasion assay, commensal bacterial strains, i.e. E. coli JCM 1649T and F. varium 113 and ATCC 8501T, induced TNF-α production by SW-480 cells. E. coli JCM 1649T stimulated the greatest TNF-α production, followed by F. varium ATCC 8501T and F. varium 113. Since the TNF-α levels at the 95 % confidence interval for the bacteria were above those of the controls, these bacteria significantly stimulated TNF-α production by SW-480 cells.
Fig. 3.
Production of cytokines upon infection with commensal bacteria and probiotics in HT-29 cells. (a) IL-8. In the invasion assay, F. varium 113 stimulated the greatest IL-8 production, followed by F. varium ATCC 8501T, E. coli JCM 1649T, B. vulgatus JCM 5826T, C. clostridioforme 94 and B. vulgatus 90. Since the IL-8 levels at the 95 % confidence interval for these bacteria were above those of the controls (<16 pg ml−1), these bacteria significantly stimulated IL-8 production by HT-29 cells. The IL-8 levels with F. varium 113 and ATCC 8501T and E. coli JCM 1649T were significantly higher than those with B. vulgatus 90 and C. clostridioforme 94 (P <0.0001). (b) TNF-α. In the invasion assay, commensal bacterial strains, i.e. F. varium 113, ATCC 8501T and E. coli JCM 1649T, induced TNF-α production by HT-29 cells. F. varium 113 stimulated the greatest TNF-α production, followed by F. varium ATCC 8501T and E. coli JCM 1649T. Since the TNF-α levels at the 95 % confidence interval for the bacteria were above those of the controls, these bacteria significantly stimulated TNF-α production by HT-29 cells.
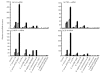
Fig. 4.
Induction of cytokine mRNAs in SW-480 cells infected with commensal bacteria and probiotics. Dotted bars, the attachment assay; black bars, the invasion assay. (a) IL-8 mRNA. In the attachment assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.05). In the invasion assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P <0.0001). The IL-8 mRNA levels with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T in the invasion assay were significantly higher than those in the attachment assay (P <0.0001). (b) TNF-α mRNA. In the attachment assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T and probiotics (P <0.0001). (c) MCP-1 mRNA. In the attachment assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T, E. coli R-1 and probiotics (P <0.0001). (d) IL-6 mRNA. In the attachment assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113 and E. coli JCM 1649T were significantly higher than those with probiotics (_P_=0.023–<0.0001). In the invasion assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (_P_=0.0081–<0.0001).
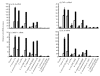
Fig. 5.
Induction of cytokine mRNAs in HT-29 cells infected with commensal bacteria and probiotics. White bars, the attachment assay; black bars, the invasion assay. (a) IL-8 mRNA. In the attachment assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.01). In the invasion assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P <0.0001). The IL-8 mRNA levels with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T in the invasion assay were significantly higher than those in the attachment assay (P <0.0001). (b) TNF-α mRNA. In the attachment assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T and probiotics (P <0.0001). (c) MCP-1 mRNA. In the attachment assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T, E. coli R-1 and probiotics (P <0.0001). (d) IL-6 mRNA. In the attachment assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113 and E. coli JCM 1649T were significantly higher than those with probiotics (_P_=0.023–<0.0001). In the invasion assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P <0.0001).
Fig. 6.
Representative photographs of immunohistochemical expression of TNF-α, IL-8, NF-_κ_B p65 and phosphor-NF-_κ_B p65 in semiserial sections of inflamed epithelium featuring cryptitis or crypt abscesses. (a) TNF-α expression in a crypt abscess. Note positive granules in the epithelial cytoplasm (arrows) (bar, 25 μm). (b) IL-8 expression in a crypt abscess. Note the positive reaction in the epithelial cytoplasm. (c) NF-_κ_B p65 expression in epithelial nuclei in a crypt abscess (arrows). (d) Phosphor-NF-_κ_B p65 expression in epithelial nuclei in a crypt abscess (arrows).
Similar articles
- Development of ulcerative colitis and its associated colorectal neoplasia as a model of the organ-specific chronic inflammation-carcinoma sequence.
Okayasu I. Okayasu I. Pathol Int. 2012 Jun;62(6):368-80. doi: 10.1111/j.1440-1827.2012.02807.x. Epub 2012 Mar 16. Pathol Int. 2012. PMID: 22612505 Review. - Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis.
Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Ohkusa T, et al. Gut. 2003 Jan;52(1):79-83. doi: 10.1136/gut.52.1.79. Gut. 2003. PMID: 12477765 Free PMC article. - Lactobacillus plantarum 299: beneficial in vitro immunomodulation in cells extracted from inflamed human colon.
Pathmakanthan S, Li CK, Cowie J, Hawkey CJ. Pathmakanthan S, et al. J Gastroenterol Hepatol. 2004 Feb;19(2):166-73. doi: 10.1111/j.1440-1746.2004.03181.x. J Gastroenterol Hepatol. 2004. PMID: 14731126
Cited by
- The role of bacteria in the pathogenesis of ulcerative colitis.
Sasaki M, Klapproth JM. Sasaki M, et al. J Signal Transduct. 2012;2012:704953. doi: 10.1155/2012/704953. Epub 2012 Apr 24. J Signal Transduct. 2012. PMID: 22619714 Free PMC article. - Periodontitis may induce gut microbiota dysbiosis via salivary microbiota.
Bao J, Li L, Zhang Y, Wang M, Chen F, Ge S, Chen B, Yan F. Bao J, et al. Int J Oral Sci. 2022 Jun 23;14(1):32. doi: 10.1038/s41368-022-00183-3. Int J Oral Sci. 2022. PMID: 35732628 Free PMC article. - Gut microbiota in pathophysiology, diagnosis, and therapeutics of inflammatory bowel disease.
Pandey H, Jain D, Tang DWT, Wong SH, Lal D. Pandey H, et al. Intest Res. 2024 Jan;22(1):15-43. doi: 10.5217/ir.2023.00080. Epub 2023 Nov 8. Intest Res. 2024. PMID: 37935653 Free PMC article. Review. - Immune reaction and colorectal cancer: friends or foes?
Formica V, Cereda V, Nardecchia A, Tesauro M, Roselli M. Formica V, et al. World J Gastroenterol. 2014 Sep 21;20(35):12407-19. doi: 10.3748/wjg.v20.i35.12407. World J Gastroenterol. 2014. PMID: 25253941 Free PMC article. Review. - Evolution of invasion in a diverse set of Fusobacterium species.
Manson McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW, Allen-Vercoe E, Earl AM. Manson McGuire A, et al. mBio. 2014 Nov 4;5(6):e01864. doi: 10.1128/mBio.01864-14. mBio. 2014. PMID: 25370491 Free PMC article.
References
- Andoh, A., Sakata, S., Koizumi, Y., Mitsuyama, K., Fujiyama, Y. & Benno, Y. (2007). Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis 13, 955–962. - PubMed
- Eckmann, L., Jung, H. C., Schurer-Maly, C., Panja, A., Morzycka-Wroblewska, E. & Kagnoff, M. F. (1993b). Differential cytokine expression by human intestinal epithelial cell lines: regulated expression of interleukin 8. Gastroenterology 105, 1689–1697. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 AI009268/AI/NIAID NIH HHS/United States
- R01 GM054666/GM/NIGMS NIH HHS/United States
- GM54666/GM/NIGMS NIH HHS/United States
- AI09268/AI/NIAID NIH HHS/United States
- DE08240/DE/NIDCR NIH HHS/United States
- DE08293/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous