Transcript length bias in RNA-seq data confounds systems biology - PubMed (original) (raw)
Transcript length bias in RNA-seq data confounds systems biology
Alicia Oshlack et al. Biol Direct. 2009.
Abstract
Background: Several recent studies have demonstrated the effectiveness of deep sequencing for transcriptome analysis (RNA-seq) in mammals. As RNA-seq becomes more affordable, whole genome transcriptional profiling is likely to become the platform of choice for species with good genomic sequences. As yet, a rigorous analysis methodology has not been developed and we are still in the stages of exploring the features of the data.
Results: We investigated the effect of transcript length bias in RNA-seq data using three different published data sets. For standard analyses using aggregated tag counts for each gene, the ability to call differentially expressed genes between samples is strongly associated with the length of the transcript.
Conclusion: Transcript length bias for calling differentially expressed genes is a general feature of current protocols for RNA-seq technology. This has implications for the ranking of differentially expressed genes, and in particular may introduce bias in gene set testing for pathway analysis and other multi-gene systems biology analyses.
Reviewers: This article was reviewed by Rohan Williams (nominated by Gavin Huttley), Nicole Cloonan (nominated by Mark Ragan) and James Bullard (nominated by Sandrine Dudoit).
Figures
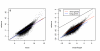
Figure 1
Differential expression as a function of transcript length. The data is binned according to transcript length and the percentage of transcripts called differentially expressed using a statistical cut-off is plotted (points). A linear regression is also plotted (lines). a – e use all the data from RNA-seq and the microarrays from studies [4-6] respectively. f and g plot 33% of genes with highest expression levels (blue crosses) and 33% of genes with low expression (red triangles) taken from the microarray data for genes which appear on both platforms in [6]. The regression gives a significant trend for the percent of differential expression with transcript length for a, c, d and f and the lowly expressed genes in g. Note that this figure illustrates common data features between disparate experiments and is not a comparison between platforms, methods or experiments.
Figure 2
Mean-variance relationship. Here we show the sample variance across lanes in the liver sample from the Marioni et al[6] data plotted as a function of the mean for each gene (a). Next we have the same data where the tag counts for each gene are divided by the length of the gene (b). The red line fits a linear relationship between the mean and variance for the one third of shortest genes while the blue line is the linear fit to the longest genes. In plot a the fits are very close to the line of equality between mean and variance (black line) which is what would be expected from a Poisson process. In plot b the short genes have higher variance for a given expression level than long genes.
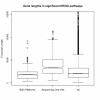
Figure 3
Length of genes found in KEGG pathways significantly over represented with differentially expressed genes. The first box in the plot represents the length of genes found in the four significant categories from both platforms. The second box is the length of genes found in categories significant only in the sequencing data. The third box is the length of all genes in common to both technologies. It can be seen that categories unique to the sequencing data tend to have longer transcripts.
Similar articles
- Length bias correction for RNA-seq data in gene set analyses.
Gao L, Fang Z, Zhang K, Zhi D, Cui X. Gao L, et al. Bioinformatics. 2011 Mar 1;27(5):662-9. doi: 10.1093/bioinformatics/btr005. Epub 2011 Jan 19. Bioinformatics. 2011. PMID: 21252076 Free PMC article. - Integrative RNA-seq and microarray data analysis reveals GC content and gene length biases in the psoriasis transcriptome.
Swindell WR, Xing X, Voorhees JJ, Elder JT, Johnston A, Gudjonsson JE. Swindell WR, et al. Physiol Genomics. 2014 Aug 1;46(15):533-46. doi: 10.1152/physiolgenomics.00022.2014. Epub 2014 May 20. Physiol Genomics. 2014. PMID: 24844236 Free PMC article. - An empirical strategy to detect bacterial transcript structure from directional RNA-seq transcriptome data.
Wang Y, MacKenzie KD, White AP. Wang Y, et al. BMC Genomics. 2015 May 7;16(1):359. doi: 10.1186/s12864-015-1555-8. BMC Genomics. 2015. PMID: 25947005 Free PMC article. - [Transcriptomes for serial analysis of gene expression].
Marti J, Piquemal D, Manchon L, Commes T. Marti J, et al. J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French. - Towards next generation CHO cell biology: Bioinformatics methods for RNA-Seq-based expression profiling.
Monger C, Kelly PS, Gallagher C, Clynes M, Barron N, Clarke C. Monger C, et al. Biotechnol J. 2015 Jul;10(7):950-66. doi: 10.1002/biot.201500107. Epub 2015 Jun 9. Biotechnol J. 2015. PMID: 26058739 Review.
Cited by
- Leveraging explainable deep learning methodologies to elucidate the biological underpinnings of Huntington's disease using single-cell RNA sequencing data.
Gao S, Wang Y, Wang J, Dong Y. Gao S, et al. BMC Genomics. 2024 Oct 4;25(1):930. doi: 10.1186/s12864-024-10855-5. BMC Genomics. 2024. PMID: 39367331 Free PMC article. - A Haloarchaeal Transcriptional Regulator That Represses the Expression of CRISPR-Associated Genes.
Turgeman-Grott I, Shalev Y, Shemesh N, Levy R, Eini I, Pasmanik-Chor M, Gophna U. Turgeman-Grott I, et al. Microorganisms. 2024 Aug 27;12(9):1772. doi: 10.3390/microorganisms12091772. Microorganisms. 2024. PMID: 39338447 Free PMC article. - Analyzing RNA-Seq data from Chlamydia with super broad transcriptomic activation: challenges, solutions, and implications for other systems.
Wan D, Cheng A, Wang Y, Zhong G, Li WV, Fan H. Wan D, et al. BMC Genomics. 2024 Aug 25;25(1):801. doi: 10.1186/s12864-024-10714-3. BMC Genomics. 2024. PMID: 39182031 Free PMC article. - Comprehensive Metatranscriptomic Analysis of Plant Viruses in Imported Frozen Cherries and Blueberries.
Lee GE, Lee HJ, Jeong RD. Lee GE, et al. Plant Pathol J. 2024 Aug;40(4):377-389. doi: 10.5423/PPJ.OA.06.2024.0088. Epub 2024 Aug 1. Plant Pathol J. 2024. PMID: 39117336 Free PMC article. - A practical introduction to holo-omics.
Odriozola I, Rasmussen JA, Gilbert MTP, Limborg MT, Alberdi A. Odriozola I, et al. Cell Rep Methods. 2024 Jul 15;4(7):100820. doi: 10.1016/j.crmeth.2024.100820. Epub 2024 Jul 9. Cell Rep Methods. 2024. PMID: 38986611 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous