Functional genomic analysis of frataxin deficiency reveals tissue-specific alterations and identifies the PPARgamma pathway as a therapeutic target in Friedreich's ataxia - PubMed (original) (raw)
. 2009 Jul 1;18(13):2452-61.
doi: 10.1093/hmg/ddp183. Epub 2009 Apr 17.
Affiliations
- PMID: 19376812
- PMCID: PMC2694693
- DOI: 10.1093/hmg/ddp183
Functional genomic analysis of frataxin deficiency reveals tissue-specific alterations and identifies the PPARgamma pathway as a therapeutic target in Friedreich's ataxia
Giovanni Coppola et al. Hum Mol Genet. 2009.
Abstract
Friedreich's ataxia (FRDA), the most common inherited ataxia, is characterized by focal neurodegeneration, diabetes mellitus and life-threatening cardiomyopathy. Frataxin, which is significantly reduced in patients with this recessive disorder, is a mitochondrial iron-binding protein, but how its deficiency leads to neurodegeneration and metabolic derangements is not known. We performed microarray analysis of heart and skeletal muscle in a mouse model of frataxin deficiency, and found molecular evidence of increased lipogenesis in skeletal muscle, and alteration of fiber-type composition in heart, consistent with insulin resistance and cardiomyopathy, respectively. Since the peroxisome proliferator-activated receptor gamma (PPARgamma) pathway is known to regulate both processes, we hypothesized that dysregulation of this pathway could play a key role in frataxin deficiency. We confirmed this by showing a coordinate dysregulation of the PPARgamma coactivator Pgc1a and transcription factor Srebp1 in cellular and animal models of frataxin deficiency, and in cells from FRDA patients, who have marked insulin resistance. Finally, we show that genetic modulation of the PPARgamma pathway affects frataxin levels in vitro, supporting PPARgamma as a novel therapeutic target in FRDA.
Figures
Figure 1.
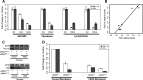
Microarray analysis of heart and skeletal muscle in KIKO mice. (A) Array clustering based on mean average deviation shows sample clustering by tissue. Within tissues, samples tend to cluster by genotype. Color coding bars: Top (genotype): red, WT samples; black, KIKO samples. Left side (tissue): red, skeletal muscle samples; black, heart samples. (B) Number of differentially expressed genes at P < 0.005. Three-hundred and twenty-one probes (194 downregulated, 127 upregulated) were differentially expressed in skeletal muscle; 174 (106 downregulated, 68 upregulated) in heart of KIKO mice versus WT; (C) Heatmap depicting fold changes in cardiac and skeletal muscle samples in KIKO mice versus controls. Shades of red represent upregulation, shades of green downregulation. Genes and samples are clustered by similarity. Most genes have tissue-specific changes, whereas a subset of genes present changes across tissues. Color coding bar: yellow, skeletal muscle samples (KIKO versus average of WT); orange: heart samples (KIKO versus average of WT); (D and E) Gene ontology analysis of differentially expressed genes in heart and skeletal muscle samples from KIKO versus WT mice. Top categories include ‘sterol’ and ‘lipid metabolic process’ in skeletal muscle and ‘regulation of muscle contraction’ in heart. Bars represent the –Log of the over-representation _P_-value, as calculated by the DAVID algorithm (
http://david.abcc.ncifcrf.gov/
). Red and green represent the proportion of up and downregulated genes, respectively. The number at the end of the bar represents the number of probes differentially expressed in each category; (F) Real-time quantitative PCR (qPCR) confirmation on 14 among the top differentially expressed genes. Fold changes are expressed in log2. Error bars: standard error. qPCR confirmed tissue specificity (e.g. Myh4 only downregulated in heart, Scd2, Slc2a5, Acly only upregulated in skeletal muscle).
Figure 2.
C2C12 (A) and HL-1 (B) cells were transfected with shRNAi for frataxin. Expression levels of frataxin versus cells transfected with control shRNAi were assessed at 12, 24, 32, 48 and 72 h after transfection (top panel). Expression levels of nine genes were assessed at 24 and 48 h after transfection (bottom panel). Red bars: upregulated genes; green bars: downregulated genes. Fold changes are expressed in log2. Error bars: standard error. Asterisk: P < 0.05, one-sample _t_-test. (C) Representative western blots (top) and blot quantification (bottom) for Frataxin and Srebp1 after transfection with control shRNAi (left) and shRNAiFxn (right). (D) shRNAiFxn transfection reduces Pgc1a expression as measured by a luciferase assay in 293 cells after 24 h. (E) FRDA patients have a significantly lower insulin sensitivity index: boxplots representing the insulin sensitivity index SI [×10−5 min−1/(μU/ml)] in healthy controls and FRDA patients, matched for age and body mass index. n = 14 in each group, **P < 0.005 by Wilcoxon rank sum test. (F) In FRDA patients, SI was significantly inversely correlated with the number of GAA repeats on the smaller FRDA allele (r = −0.55, P < 0.05). (G) Physical impairment scores are not significantly different between insulin-resistant and insulin-sensitive patients; the bar graph depicts the SI (left) and the physical impairment scores [on a scale from 0 (normal) to 42 (most severe impairment), right] of the 50% (n = 7) most insulin sensitive (light gray bars) and of the 50% (n = 7) most insulin resistant patients (dark gray bars); P = 0.5 for comparison between the two groups.
Figure 3.
Genetic and pharmacologic modulation of the PPARγ pathway affects frataxin levels in vitro. (A and B) Pgc1a levels are correlated with frataxin levels in neural precursor cells from mouse models and in cells from patients with FRDA. (A) qPCR quantification of F_xn_ (white bars) and Pgc1a (blue bars) in neural precursor cells from the subventricular zone (SVZ-NPC) in wild-type (WT, n = 3), KIKI (n = 3) and KIKO (n = 3) mice, and in lymphoblasts and fibroblasts from FRDA patients (n = 4) and normal controls (n = 3). KIKI mice express ∼75% of normal frataxin levels, and KIKO ∼30%. Pgc1a mRNA expression levels are reduced in all these cell lines, in a degree which is proportional to Fxn downregulation. Bars represent the average of six replicates, error bars represent the standard error. P < 0.05 for all the comparisons; (B) Relative Pgc1a levels are strongly correlated with relative Fxn mRNA levels (_r_2 = 0.9, P < 0.001); qPCR quantification in fibroblasts from controls (open circles, n = 3) and FRDA patients (filled circles, n = 4). (C and D) PGC1A downregulation via siRNAi reduces frataxin protein levels in control fibroblasts, and further in FRDA fibroblasts. Western blotting analysis (C) and quantification (D) of PGC-1A (top) and FXN (bottom) protein 72 h after transfection with siRNAiPgc1a. Control and FRDA are fibroblasts from healthy controls and patients, respectively. All experiments were performed on n = 4 FRDA cell lines and n = 3 control cell lines. Bars represent the average of six replicates, error bars represent the standard error. P < 0.05 for all comparisons.
Figure 4.
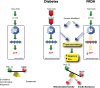
Diabetes in FRDA and type-2 diabetes may have a final common pathway. In normal conditions (left panel), lipid breakdown and biosynthesis are tightly controlled by the two master regulators Pgc-1a and Srebp-1. In type-2 diabetes (center), increased dietary lipids provided to the skeletal muscle can overload TCA and divert long-chain-CoA species into lipid precursors than can be converted in lipids, leading to an increased lipid intramyocellular content, which in turn leads to reduced glucose transport, reduced glycogen synthesis and ultimately to insulin resistance. In FRDA (right), deficits of TCA enzymes occur, as well as decreased activity of complexes I–II–III of the respiratory chain. An ineffective TCA and mitochondrial OXPHOS would lead to TCA overflow, favoring a metabolic shift toward the redirection of citrate from the TCA cycle and electron transport chain to processing into fatty acids. In mitochondrial diseases and in FRDA in particular, the oxidative stress can contribute to the diabetes pathogenesis at both (i) skeletal muscle and liver (insulin targets) and (ii) the beta-cell level, leading to overt diabetes mellitus. TCA, tricarboxylic acid cycle; IMLs, intra-myocellular lipids.
Comment in
- Diverse effects in Friedreich's ataxia place PGC-1alpha center-stage.
Metzler M. Metzler M. Clin Genet. 2009 Oct;76(4):345-7. doi: 10.1111/j.1399-0004.2009.01260_3.x. Clin Genet. 2009. PMID: 19793309 No abstract available.
Similar articles
- PPAR gamma agonist leriglitazone improves frataxin-loss impairments in cellular and animal models of Friedreich Ataxia.
Rodríguez-Pascau L, Britti E, Calap-Quintana P, Dong YN, Vergara C, Delaspre F, Medina-Carbonero M, Tamarit J, Pallardó FV, Gonzalez-Cabo P, Ros J, Lynch DR, Martinell M, Pizcueta P. Rodríguez-Pascau L, et al. Neurobiol Dis. 2021 Jan;148:105162. doi: 10.1016/j.nbd.2020.105162. Epub 2020 Nov 7. Neurobiol Dis. 2021. PMID: 33171227 - PPAR-gamma agonist Azelaoyl PAF increases frataxin protein and mRNA expression: new implications for the Friedreich's ataxia therapy.
Marmolino D, Acquaviva F, Pinelli M, Monticelli A, Castaldo I, Filla A, Cocozza S. Marmolino D, et al. Cerebellum. 2009 Jun;8(2):98-103. doi: 10.1007/s12311-008-0087-z. Epub 2008 Dec 23. Cerebellum. 2009. PMID: 19104905 - Elucidation of the mechanism of mitochondrial iron loading in Friedreich's ataxia by analysis of a mouse mutant.
Huang ML, Becker EM, Whitnall M, Suryo Rahmanto Y, Ponka P, Richardson DR. Huang ML, et al. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16381-6. doi: 10.1073/pnas.0906784106. Epub 2009 Sep 4. Proc Natl Acad Sci U S A. 2009. PMID: 19805308 Free PMC article. - Neurodegeneration in Friedreich's ataxia: from defective frataxin to oxidative stress.
Gomes CM, Santos R. Gomes CM, et al. Oxid Med Cell Longev. 2013;2013:487534. doi: 10.1155/2013/487534. Epub 2013 Jul 9. Oxid Med Cell Longev. 2013. PMID: 23936609 Free PMC article. Review. - Therapeutic Prospects for Friedreich's Ataxia.
Zhang S, Napierala M, Napierala JS. Zhang S, et al. Trends Pharmacol Sci. 2019 Apr;40(4):229-233. doi: 10.1016/j.tips.2019.02.001. Trends Pharmacol Sci. 2019. PMID: 30905359 Free PMC article. Review.
Cited by
- Iron homeostasis and ferroptosis in human diseases: mechanisms and therapeutic prospects.
Ru Q, Li Y, Chen L, Wu Y, Min J, Wang F. Ru Q, et al. Signal Transduct Target Ther. 2024 Oct 14;9(1):271. doi: 10.1038/s41392-024-01969-z. Signal Transduct Target Ther. 2024. PMID: 39396974 Free PMC article. Review. - Skeletal Muscle Involvement in Friedreich Ataxia.
Indelicato E, Wanschitz J, Löscher W, Boesch S. Indelicato E, et al. Int J Mol Sci. 2024 Sep 13;25(18):9915. doi: 10.3390/ijms25189915. Int J Mol Sci. 2024. PMID: 39337401 Free PMC article. Review. - Interplay of FXN expression and lipolysis in white adipocytes plays a critical role in insulin sensitivity in Friedreich's ataxia mouse model.
Wu L, Huang F, Yang L, Yang L, Sun Z, Zhang J, Xia S, Zhao H, Ding Y, Bian D, Li K. Wu L, et al. Sci Rep. 2024 Aug 27;14(1):19876. doi: 10.1038/s41598-024-71099-7. Sci Rep. 2024. PMID: 39191875 Free PMC article. - New and Emerging Drug and Gene Therapies for Friedreich Ataxia.
Scott V, Delatycki MB, Tai G, Corben LA. Scott V, et al. CNS Drugs. 2024 Oct;38(10):791-805. doi: 10.1007/s40263-024-01113-z. Epub 2024 Aug 8. CNS Drugs. 2024. PMID: 39115603 Free PMC article. Review. - Emerging antioxidant therapies in Friedreich's ataxia.
Edzeamey FJ, Ramchunder Z, Pourzand C, Anjomani Virmouni S. Edzeamey FJ, et al. Front Pharmacol. 2024 Feb 6;15:1359618. doi: 10.3389/fphar.2024.1359618. eCollection 2024. Front Pharmacol. 2024. PMID: 38379897 Free PMC article. Review.
References
- Pandolfo M. Friedreich ataxia. Semin. Pediatr. Neurol. 2003;10:163–172. - PubMed
- Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. - PubMed
- Puccio H., Simon D., Cossee M., Filipe C., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. - PubMed
- Thierbach R., Schulz T.J., Isken F., Voigt A., Mietzner B., Drewes G., von Kleist-Retzow J.C., Wiesner R.J., Magnuson M.A., Puccio H., et al. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Hum. Mol. Genet. 2005;14:3857–3864. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases