FastTree: computing large minimum evolution trees with profiles instead of a distance matrix - PubMed (original) (raw)
FastTree: computing large minimum evolution trees with profiles instead of a distance matrix
Morgan N Price et al. Mol Biol Evol. 2009 Jul.
Abstract
Gene families are growing rapidly, but standard methods for inferring phylogenies do not scale to alignments with over 10,000 sequences. We present FastTree, a method for constructing large phylogenies and for estimating their reliability. Instead of storing a distance matrix, FastTree stores sequence profiles of internal nodes in the tree. FastTree uses these profiles to implement Neighbor-Joining and uses heuristics to quickly identify candidate joins. FastTree then uses nearest neighbor interchanges to reduce the length of the tree. For an alignment with N sequences, L sites, and a different characters, a distance matrix requires O(N(2)) space and O(N(2)L) time, but FastTree requires just O(NLa + N ) memory and O(N log (N)La) time. To estimate the tree's reliability, FastTree uses local bootstrapping, which gives another 100-fold speedup over a distance matrix. For example, FastTree computed a tree and support values for 158,022 distinct 16S ribosomal RNAs in 17 h and 2.4 GB of memory. Just computing pairwise Jukes-Cantor distances and storing them, without inferring a tree or bootstrapping, would require 17 h and 50 GB of memory. In simulations, FastTree was slightly more accurate than Neighbor-Joining, BIONJ, or FastME; on genuine alignments, FastTree's topologies had higher likelihoods. FastTree is available at http://microbesonline.org/fasttree.
Figures
FIG. 1.—
Overview of FastTree.
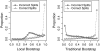
FIG. 2.—
Distribution of support values for simulated alignments of 250 protein sequences with gaps. We compare the distribution of FastTree's local bootstrap and the traditional (global) bootstrap for correctly and incorrectly inferred splits. The right-most bin contains the strongly supported splits (0.95–1.0)
Similar articles
- FastTree 2--approximately maximum-likelihood trees for large alignments.
Price MN, Dehal PS, Arkin AP. Price MN, et al. PLoS One. 2010 Mar 10;5(3):e9490. doi: 10.1371/journal.pone.0009490. PLoS One. 2010. PMID: 20224823 Free PMC article. - RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation.
Liu K, Linder CR, Warnow T. Liu K, et al. PLoS One. 2011;6(11):e27731. doi: 10.1371/journal.pone.0027731. Epub 2011 Nov 21. PLoS One. 2011. PMID: 22132132 Free PMC article. - Assessment of protein distance measures and tree-building methods for phylogenetic tree reconstruction.
Hollich V, Milchert L, Arvestad L, Sonnhammer EL. Hollich V, et al. Mol Biol Evol. 2005 Nov;22(11):2257-64. doi: 10.1093/molbev/msi224. Epub 2005 Jul 27. Mol Biol Evol. 2005. PMID: 16049194 - Genome BLAST distance phylogenies inferred from whole plastid and whole mitochondrion genome sequences.
Auch AF, Henz SR, Holland BR, Göker M. Auch AF, et al. BMC Bioinformatics. 2006 Jul 19;7:350. doi: 10.1186/1471-2105-7-350. BMC Bioinformatics. 2006. PMID: 16854218 Free PMC article.
Cited by
- A genetic barcode of SARS-CoV-2 for monitoring global distribution of different clades during the COVID-19 pandemic.
Guan Q, Sadykov M, Mfarrej S, Hala S, Naeem R, Nugmanova R, Al-Omari A, Salih S, Al Mutair A, Carr MJ, Hall WW, Arold ST, Pain A. Guan Q, et al. Int J Infect Dis. 2020 Nov;100:216-223. doi: 10.1016/j.ijid.2020.08.052. Epub 2020 Aug 22. Int J Infect Dis. 2020. PMID: 32841689 Free PMC article. - Role and Modulation of TRPV1 in Mammalian Spermatozoa: An Updated Review.
Ramal-Sanchez M, Bernabò N, Valbonetti L, Cimini C, Taraschi A, Capacchietti G, Machado-Simoes J, Barboni B. Ramal-Sanchez M, et al. Int J Mol Sci. 2021 Apr 21;22(9):4306. doi: 10.3390/ijms22094306. Int J Mol Sci. 2021. PMID: 33919147 Free PMC article. Review. - The genome of a hadal sea cucumber reveals novel adaptive strategies to deep-sea environments.
Shao G, He T, Mu Y, Mu P, Ao J, Lin X, Ruan L, Wang Y, Gao Y, Liu D, Zhang L, Chen X. Shao G, et al. iScience. 2022 Nov 9;25(12):105545. doi: 10.1016/j.isci.2022.105545. eCollection 2022 Dec 22. iScience. 2022. PMID: 36444293 Free PMC article. - Novel transglutaminase-like peptidase and C2 domains elucidate the structure, biogenesis and evolution of the ciliary compartment.
Zhang D, Aravind L. Zhang D, et al. Cell Cycle. 2012 Oct 15;11(20):3861-75. doi: 10.4161/cc.22068. Epub 2012 Sep 14. Cell Cycle. 2012. PMID: 22983010 Free PMC article.
References
- Bininda-Emonds OR, Brady SG, Kim J, Sanderson MJ. Scaling of accuracy in extremely large phylogenetic trees. Pac Symp Biocomput. 2001;2001:547–558. - PubMed
- DeLong ER, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1998;44:837–845. - PubMed
- Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol. 2002;9:687–705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources