Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection - PubMed (original) (raw)
Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection
Amy Croswell et al. Infect Immun. 2009 Jul.
Abstract
The impact of antibiotics on the host's protective microbiota and the resulting increased susceptibility to mucosal infection are poorly understood. In this study, antibiotic regimens commonly applied to murine enteritis models are used to examine the impact of antibiotics on the intestinal microbiota, the time course of recovery of the biota, and the resulting susceptibility to enteric Salmonella infection. Molecular analysis of the microbiota showed that antibiotic treatment has an impact on the colonization of the murine gut that is site and antibiotic dependent. While combinations of antibiotics were able to eliminate culturable bacteria, none of the antibiotic treatments were effective at sterilizing the intestinal tract. Recovery of total bacterial numbers occurs within 1 week after antibiotic withdrawal, but alterations in specific bacterial groups persist for several weeks. Increased Salmonella translocation associated with antibiotic pretreatment corrects rapidly in association with the recovery of the most dominant bacterial group, which parallels the recovery of total bacterial numbers. However, susceptibility to intestinal colonization and mucosal inflammation persists when mice are infected several weeks after withdrawal of antibiotics, correlating with subtle alterations in the intestinal microbiome involving alterations of specific bacterial groups. These results show that the colonizing microbiotas are integral to mucosal host protection, that specific features of the microbiome impact different aspects of enteric Salmonella pathogenesis, and that antibiotics can have prolonged deleterious effects on intestinal colonization resistance.
Figures
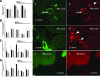
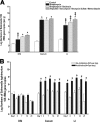
FIG. 1.
Impact of antibiotic regimens on intestinal bacterial colonization. Five-week-old female mice were given antibiotics, as indicated, in their drinking water for 7 days (n = 5 mice per group). Total bacterial genomic DNA was isolated from the DSI, cecum, and LI of each mouse and analyzed by qPCR for total bacterial numbers (A) and the abundance of specific bacterial groups (per gram) in the in the DSI (B), cecum (C), and LI (D). In the DSI, cecum, and LI, antibiotic treatment groups are all significantly different from untreated controls for E. rectale-C. coccoides (Erec), Lactobacillus (Lact), and SFB (P < 0.05). * indicates statistically significant differences from the control mice (P < 0.05); ‡ indicates statistically significant differences from all other treatment groups (P < 0.05). (E) FISH using a mixture of a Texas Red universal bacterial probe to show all bacteria (b, d, and f) and a FAM-SFB-specific bacterial probe to show SFB (a, c, and e) was performed on the terminal ileum of control (a and b), streptomycin-treated (c and d), and streptomycin-bacitracin-treated (e and f) mice and visualized by fluorescence microscopy. These images are representative of each animal studied (n = 5 mice per group). Arrows point to SFB. Arrowheads point to non-SFB commensal bacteria. The mucosal surface and lumen are labeled.
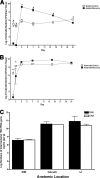
FIG. 2.
Time course of recovery of intestinal colonization. (A and B) Parallel groups of mice (n = 5 mice per group) were given untreated drinking water (open symbols) or bacitracin-streptomycin in drinking water (filled symbols) for 1 week. The antibiotics were withdrawn, and the biome was allowed to recover for 1, 3, 7, 14, or 21 days. At each time point, the LI bacteria were cultured for aerobic (A) and anaerobic (B) bacteria. * indicates statistically significant differences from the control mice (P < 0.001). To determine whether qPCR from genomic DNA represented live bacteria in the GI tract, analysis and quantification of 16S copies were performed and compared for bacterial DNA and RNA. Total bacterial RNA and genomic DNA were isolated from the DSI, cecum, and LI of adult unchallenged mice (n = 3). (C) Total 16S gene copies were quantified from genomic DNA (white bars) by qPCR and from RNA (black bars) by RT-qPCR.
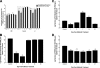
FIG. 3.
Incomplete recovery of the intestinal microbiome after antibiotic treatment. Parallel groups of mice (n = 5 mice per group) were given untreated drinking water or bacitracin-streptomycin in drinking water for 1 week. The antibiotics were withdrawn, and the biome was allowed to recover for 1, 3, 7, 14, or 21 days. Total bacterial genomic DNA was isolated from the DSI, cecum, and LI of each mouse. qPCR was performed to quantify numbers of total bacteria (A), the E. rectale-C. coccoides group (Erec) (B), the Enterobacteriaceae (Ent) (C), and SFB (D) per gram. * indicates statistically significant differences from control mice (P < 0.001).
FIG. 4.
Effect of antibiotic regimens on Salmonella translocation. Parallel groups of mice (n = 4 mice per group) were given untreated drinking water or the indicated regimens of antibiotics for 1 week. The antibiotics were withdrawn, and the mice were inoculated with Salmonella (108 CFU/mouse) by intragastric gavage and sacrificed after 3 days. (A) The livers were isolated, homogenized, and analyzed by dilution plating onto selective agar to determine the bacterial burden and extent of translocation. Each symbol represents one mouse. The horizontal bars indicate the means of data from the four mice per group (P = 0.0055 by Wilcoxon rank-sum test). Data for all antibiotic treatment groups are significantly different from those of the controls. Tukey's multiple comparison shows that the values for antibiotic treatment groups are significantly greater than those for the control group, but they are not significantly different from each other. (B) Parallel groups of mice (n = 5 mice per group) were given untreated drinking water or bacitracin-streptomycin (Strep/Bac) in drinking water for 1 week. The antibiotics were withdrawn, and the biome was allowed to recover for 1, 3, 7, 14, or 21 days before oral infection with 108 CFU of Salmonella (open squares). Mice that were not treated with antibiotics were used for control infections (closed squares). Salmonella translocation into the liver (per gram) was determined 3 days postinoculation by dilution plating onto selective agar. Each symbol represents one mouse. The horizontal bar indicates the mean data from five mice per group. * indicates statistically significant differences (P < 0.05). PT, posttreatment.
FIG. 5.
Antibiotic treatment enhances Salmonella colonization of the intestinal tract. (A) Mice (n = 5 mice per group) were treated with different regimens of antibiotics for 1 week, followed by Salmonella inoculation by intragastric gavage, and sacrificed after 3 days. Total bacterial genomic DNA was isolated from the DSI, cecum, and LI and analyzed for total Salmonella burden by qPCR. * indicates statistically significant differences from control mice (P < 0.05); ‡ indicates statistically significant differences from all other treatment groups (P < 0.05). (B) Untreated control mice (white bars) and antibiotic-treated mice (black bars) were allowed to recover from treatment with bacitracin-streptomycin for 1, 3, 7, 14, or 21 days prior to challenge with Salmonella by oral gavage. Mice were sacrificed after 3 days, and total bacterial genomic DNA was isolated from each segment of the GI tract and analyzed for total Salmonella burden (per gram) by qPCR (n = 5 mice per group). * indicates statistically significant differences from control mice (P < 0.05). In the cecum and LI, there were no statistically significant differences in Salmonella colonization among antibiotic-treated mice at any time point.
FIG. 6.
Antibiotic pretreatment enhances the development of acute colitis. Mice (n = 5 mice per group) were treated with different regimens of antibiotics for 1 week, followed by Salmonella inoculation by oral gavage, and were sacrificed after 3 days. Hematoxylin- and eosin-stained sections of intestinal tissue from the DSI, cecum, and LI of representative mice were examined for evidence of inflammation and colitis. All images were taken at a ×20 magnification. Arrows point to areas of inflammation with neutrophil infiltrates.
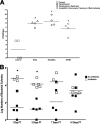
FIG. 7.
Persistent susceptibility to colitis during antibiotic recovery. Mice (n = 5 mice per group) were allowed to recover from treatment with bacitracin-streptomycin (open squares) for 1, 3, 7, 14, or 21 days prior to challenge with Salmonella by oral gavage and were sacrificed after 3 days. Control mice were not pretreated with antibiotics (closed squares). Hematoxylin- and eosin-stained sections of intestinal tissue from the DSI, cecum, and LI of each mouse (n = 5) were examined for evidence of inflammation and colitis. Cecal sections were blindly scored using histological criteria for the extent of epithelial hyperplasia, edema, and acute inflammation.
Similar articles
- Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract.
Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Barman M, et al. Infect Immun. 2008 Mar;76(3):907-15. doi: 10.1128/IAI.01432-07. Epub 2007 Dec 26. Infect Immun. 2008. PMID: 18160481 Free PMC article. - Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection.
Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Sekirov I, et al. Infect Immun. 2008 Oct;76(10):4726-36. doi: 10.1128/IAI.00319-08. Epub 2008 Aug 4. Infect Immun. 2008. PMID: 18678663 Free PMC article. - Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection.
Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. Garner CD, et al. Infect Immun. 2009 Jul;77(7):2691-702. doi: 10.1128/IAI.01570-08. Epub 2009 May 11. Infect Immun. 2009. PMID: 19433544 Free PMC article. - Antibiotics and the Intestinal Microbiome : Individual Responses, Resilience of the Ecosystem, and the Susceptibility to Infections.
Thiemann S, Smit N, Strowig T. Thiemann S, et al. Curr Top Microbiol Immunol. 2016;398:123-146. doi: 10.1007/82_2016_504. Curr Top Microbiol Immunol. 2016. PMID: 27738912 Review. - A comparative analysis of the effect of antibiotic treatment and enteric infection on intestinal homeostasis.
Antunes LC, Finlay BB. Antunes LC, et al. Gut Microbes. 2011 Mar-Apr;2(2):105-8. doi: 10.4161/gmic.2.2.15610. Epub 2011 Mar 1. Gut Microbes. 2011. PMID: 21637027 Review.
Cited by
- Gut immune maturation depends on colonization with a host-specific microbiota.
Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Chung H, et al. Cell. 2012 Jun 22;149(7):1578-93. doi: 10.1016/j.cell.2012.04.037. Cell. 2012. PMID: 22726443 Free PMC article. - Depletion of gut microbiota is associated with improved neurologic outcome following traumatic brain injury.
Simon DW, Rogers MB, Gao Y, Vincent G, Firek BA, Janesko-Feldman K, Vagni V, Kochanek PM, Ozolek JA, Mollen KP, Clark RSB, Morowitz MJ. Simon DW, et al. Brain Res. 2020 Nov 15;1747:147056. doi: 10.1016/j.brainres.2020.147056. Epub 2020 Aug 13. Brain Res. 2020. PMID: 32798452 Free PMC article. - Mouse intestinal microbiota reduction favors local intestinal immunity triggered by antigens displayed in Bacillus subtilis biofilm.
Vogt CM, Hilbe M, Ackermann M, Aguilar C, Eichwald C. Vogt CM, et al. Microb Cell Fact. 2018 Nov 26;17(1):187. doi: 10.1186/s12934-018-1030-8. Microb Cell Fact. 2018. PMID: 30477481 Free PMC article. - Evolution of the murine gut resistome following broad-spectrum antibiotic treatment.
de Nies L, Busi SB, Tsenkova M, Halder R, Letellier E, Wilmes P. de Nies L, et al. Nat Commun. 2022 Apr 28;13(1):2296. doi: 10.1038/s41467-022-29919-9. Nat Commun. 2022. PMID: 35484157 Free PMC article. - Metagenomic systems biology and metabolic modeling of the human microbiome: from species composition to community assembly rules.
Levy R, Borenstein E. Levy R, et al. Gut Microbes. 2014 Mar-Apr;5(2):265-70. doi: 10.4161/gmic.28261. Epub 2014 Feb 20. Gut Microbes. 2014. PMID: 24637600 Free PMC article.
References
- Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 3071915-1920. - PubMed
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55541-555. - PubMed
- Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI057757/AI/NIAID NIH HHS/United States
- R01 AI057757-05/AI/NIAID NIH HHS/United States
- R21 AI057757/AI/NIAID NIH HHS/United States
- AI057757/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical