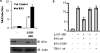Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter - PubMed (original) (raw)
. 2009 Jun;29(12):3401-12.
doi: 10.1128/MCB.00880-08. Epub 2009 Apr 20.
Myriam Vilasco, Meztli Arguello, Qiang Sun, Judith Lacoste, Thi Lien-Anh Nguyen, Tiejun Zhao, Elena A Shestakova, Scott Zaari, Annie Bibeau-Poirier, Marc J Servant, Rongtuan Lin, Eliane F Meurs, John Hiscott
Affiliations
- PMID: 19380491
- PMCID: PMC2698723
- DOI: 10.1128/MCB.00880-08
Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter
Suzanne Paz et al. Mol Cell Biol. 2009 Jun.
Abstract
Induction of the antiviral interferon response is initiated upon recognition of viral RNA structures by the RIG-I or Mda-5 DEX(D/H) helicases. A complex signaling cascade then converges at the mitochondrial adapter MAVS, culminating in the activation of the IRF and NF-kappaB transcription factors and the induction of interferon gene expression. We have previously shown that MAVS recruits IkappaB kinase epsilon (IKKepsilon) but not TBK-1 to the mitochondria following viral infection. Here we map the interaction of MAVS and IKKepsilon to the C-terminal region of MAVS and demonstrate that this interaction is ubiquitin dependent. MAVS is ubiquitinated following Sendai virus infection, and K63-linked ubiquitination of lysine 500 (K500) of MAVS mediates recruitment of IKKepsilon to the mitochondria. Real-time PCR analysis reveals that a K500R mutant of MAVS increases the mRNA level of several interferon-stimulated genes and correlates with increased NF-kappaB activation. Thus, recruitment of IKKepsilon to the mitochondria upon MAVS K500 ubiquitination plays a modulatory role in the cascade leading to NF-kappaB activation and expression of inflammatory and antiviral genes. These results provide further support for the differential role of IKKepsilon and TBK-1 in the RIG-I/Mda5 pathway.
Figures
FIG. 1.
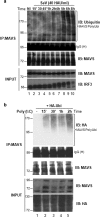
MAVS directly recruits IKKɛ to the mitochondria. (a) COS-7 cells were transfected with IKKɛ and MAVS expression plasmids. The mitochondrial network was stained with MitoTracker (blue). Cells were fixed and stained for either MAVS (red) or IKKɛ (green). The confocal images were merged. A zoom of the merge can also be seen. (b) Subcellular localization of IKKɛ and TBK-1 following SeV infection of A549 cells (4 h; 40 HAU/ml). Mitochondrial and cytoplasmic fractions were isolated and analyzed by immunoblotting with anti-IKKɛ and anti-TBK-1 antibodies. HSP70 was used as a mitochondrial marker, and β-actin was used a cytoplasmic marker. (c) HEK293 cells expressing Myc-MAVS, Flag-IKKɛ, and Flag-TBK-1 expression plasmids were submitted to immunoprecipitation with an anti-Myc antibody followed by immunoblotting with anti-FLAG antibody (top panel). Equal amounts of immunoprecipitated MAVS were revealed by immunoblotting with anti-Myc (second panel). Inputs for IKKɛ, TBK-1, and MAVS are shown. (d) Endogenous interaction between MAVS and IKKɛ was determined in A549 cells infected with SeV (40 HAU/ml) for the indicated times. Endogenous MAVS was immunoprecipitated using an anti-MAVS antibody cocktail, and interaction with IKKɛ was revealed using an anti-IKKɛ antibody (top panel). Immunoblotting with a TBK-1-specific antibody showed no interaction between MAVS and TBK-1 (second panel). Equal amounts of immunoprecipitated MAVS and IgG heavy chain are shown. Input for IKKɛ, TBK-1, and MAVS are shown (last three panels). (e) The dependence of the MAVS-IKKɛ interaction on ubiquitination was demonstrated in HEK293 cells transfected with plasmids expressing Flag-IKKɛ, HA-ubiquitin, and HA-ubiquitin-KO as indicated. Immunoprecipitation of endogenous MAVS using an anti-MAVS antibody cocktail was followed by immunoblotting with anti-Flag antibody to reveal the interaction with Flag-IKKɛ.
FIG. 2.
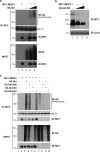
Endogenous MAVS is ubiquitinated following viral infection. (a) A549 cells were challenged with SeV (40 HAU/ml) for the indicated times followed by immunoprecipitation of endogenous MAVS with anti-MAVS cocktail. To remove possible interacting proteins, extracts were boiled in 1% SDS prior to immunoprecipitation. Immunoblotting with antiubiquitin antibody revealed a high-molecular-weight smear characteristic of ubiquitination (top panel). Equal amounts of immunoprecipitated MAVS are shown. Immunoblotting for IRF-3 phosphorylation was used to monitor activation of the IFN pathway (bottom panel). (b) Ubiquitination of MAVS following double-stranded DNA stimulation was assessed in HEK293 cells expressing HA-ubiquitin. Cells were challenged with poly(I·C) for the indicated times, and endogenous MAVS was immunoprecipitated with anti-MAVS cocktail. To remove possible interacting proteins, extracts were boiled in 1% SDS prior to immunoprecipitation. Ubiquitination signal was revealed by immunoblotting with anti-HA (top panel). Equal amounts of immunoprecipitated MAVS are shown.
FIG. 3.
MAVS undergoes K63- and K48-linked ubiquitination in vivo. (a) HEK293 cells were transfected with Myc-MAVS and increasing amounts of HA-ubiquitin as indicated. Whole-cell extracts were denatured and immunoprecipitated using an anti-Myc antibody. Immunoblotting with anti-HA revealed a high-molecular-weight smear characteristic of ubiquitination (top panel). Levels of immunoprecipitated MAVS and IgG heavy chain are shown. (b) HEK293 cells were transfected with Myc-MAVS and increasing amounts of HA-Ubi-KO as indicated. Immunoblotting with anti-MAVS antibody (N terminal) revealed unmodified MAVS protein and polyubiquitinated forms of MAVS that disappeared in the presence of Ubi-KO (top panel). Actin was used as a loading control (bottom panel). (c) HEK293 cells were transfected with Myc-MAVS, HA-Ubi, HA-Ubi-K48, HA-Ubi-KO, and HA-Ubi-K63 as indicated. Polyubiquitination of MAVS was detected as described for panel a.
FIG. 4.
IKKɛ interacts with the C terminus of MAVS and requires the TM domain. (a) Schematic representation of full-length MAVS (FL) and deletion constructs. The location of the CARD, proline-rich region (Pro), and TM domain are shown. Lysine residues of MAVS are also shown. (b) HEK293 cells were transfected with the indicated MAVS constructs. Coimmunoprecipitation was performed using an anti-Myc antibody (MAVS) followed by immunoblotting with an anti-Flag (IKKɛ) antibody (top panel). Immunoprecipitated MAVS constructs were revealed by immunoblotting with anti-Myc antibody (second panel). Equal input for Flag-IKKɛ is shown (bottom panel). (c) Cells were transfected with Flag-IKKɛ along with either wild-type MAVS full-length (FL), wild-type (468-540) truncation construct, or with mutants with a K500R substitution [FL-K500R or (468-540)-K500R]. A MAVS K136R mutant (FL-K136R) was used as a control. Coimmunoprecipitation experiments using an anti-Myc antibody followed by immunoblotting with anti-FLAG revealed lack of interaction with IKKɛ for the K500R mutants (top panel). (d) Subcellular localization of MAVS and IKKɛ following overexpression of Myc-MAVS wt, Myc-MAVS-K500R, and Flag-IKKɛ. Mitochondrial and cytoplasmic fractions were isolated and analyzed by immunoblotting with anti-Flag and anti-Myc antibodies. Tubulin was used as a cytoplasmic marker.
FIG. 5.
Lysine 500 of MAVS is an acceptor site for K63-linked ubiquitination. (a and b) HEK293 cells were transfected with the indicated plasmids. Ubiquitination of MAVS was revealed by immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-HA antibody (top panel). An equal amount of immunoprecipitated MAVS protein was observed (second panel). The IgG heavy chain is indicated. (c) HEK293 cells were transfected with either a Flag-IKKɛ or Flag-TBK-1 expression construct. Immunoprecipitation of either Flag-IKKɛ or Flag-TBK-1 was followed by incubation with a K63-ubiquitin polymer (Ub2-7) in increasing amounts. Poly-K63 linkage was revealed by immunoblotting with antiubiquitin antibody (top panel). An equal amount of either immunoprecipitated IKKɛ or TBK-1 protein is shown (second panel).
FIG. 6.
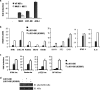
MAVS(Δ101-480) is functional but requires endogenous MAVS. (a) MAVS wt and MAVS−/− MEFs were transfected with a IFN-β-Luc reporter plasmid, and expression plasmids encoding either empty, MAVS wt, MAVS K500R, MAVS(Δ101-480), or MAVS(Δ101-480)-K500R as indicated. Luciferase activity was analyzed at 24 h posttransfection and activation was determined and compared to that with the empty vector; values represent averages ± standard deviations. Results are representative of at least three experiments run in triplicate. K500 ubiquitination negatively regulates ISG expression by affecting NF-κB signaling. (b) Quantitative PCR analysis of total RNA isolated from HeLa cells transfected with either empty vector, MAVS(Δ101-480), or MAVS(Δ101-480)-K500R. Relative expression levels of IFN-β, CXCL10, RANTES, IL-6, ISG15, ISG56, ADAR-1, IFIT3, OAS1 or STAT-1 versus GAPDH mRNA are shown. Data are representative of at least two experiments run in duplicate. (c) HeLa cells were transfected with IFN-β-Luc, RANTES-Luc, NF-κB-Luc, and p2(2)-Luc reporter plasmids and expression plasmids encoding MAVS(Δ101-480) or MAVS(Δ101-480)-K500R, as indicated. Luciferase activity was analyzed at 24 h posttransfection and activation was determined and compared to that with the empty vector; values represent the averages ± standard deviations. Results are representative of at least three experiments run in triplicate. (d) HeLa cells were transfected with either empty vector, MAVS(Δ101-480), or MAS(Δ101-480)-K500R. WCE were resolved on a 10% SDS-PAGE and transferred to a nitrocellulose membrane, and IκBα phosphorylation was detected using a phospho-specific antibody. Total IκBα levels were also monitored.
FIG. 7.
IKKɛ is required to mediate the negative effect of K500. (a) HeLa cells were transfected with control siRNA or siRNA against IKKɛ. At 48 h posttransfection, IFN-β-Luc reporter plasmid and expression plasmids encoding MAVS(Δ101-480) were also transfected. Luciferase activity was analyzed at 24 h posttransfection and activation was determined and compared to that for the empty vector; values represent the averages ± standard deviations. Results are representative of at least three experiments run in triplicate. Samples were also analyzed by immunoblotting for IKKɛ extinction by using an anti-IKKɛ antibody. Actin immunoblotting results are shown. (b) HeLa cells were transfected with an IFN-β-Luc reporter plasmid or expression plasmids encoding MAVS(Δ101-480), IKKɛ wt, IKKɛK38A, or TBK-1, as indicated. Luciferase activity was analyzed at 24 h posttransfection and activation was determined and compared to that for the empty vector; values represent the averages ± standard deviations. Results are representative of at least three experiments run in triplicate.
Similar articles
- DDX3 directly regulates TRAF3 ubiquitination and acts as a scaffold to co-ordinate assembly of signalling complexes downstream from MAVS.
Gu L, Fullam A, McCormack N, Höhn Y, Schröder M. Gu L, et al. Biochem J. 2017 Feb 15;474(4):571-587. doi: 10.1042/BCJ20160956. Epub 2016 Dec 15. Biochem J. 2017. PMID: 27980081 - Lysine 63-linked TANK-binding kinase 1 ubiquitination by mindbomb E3 ubiquitin protein ligase 2 is mediated by the mitochondrial antiviral signaling protein.
Ye JS, Kim N, Lee KJ, Nam YR, Lee U, Joo CH. Ye JS, et al. J Virol. 2014 Nov;88(21):12765-76. doi: 10.1128/JVI.02037-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142606 Free PMC article. - TRAF5 is a downstream target of MAVS in antiviral innate immune signaling.
Tang ED, Wang CY. Tang ED, et al. PLoS One. 2010 Feb 11;5(2):e9172. doi: 10.1371/journal.pone.0009172. PLoS One. 2010. PMID: 20161788 Free PMC article. - Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter.
Belgnaoui SM, Paz S, Hiscott J. Belgnaoui SM, et al. Curr Opin Immunol. 2011 Oct;23(5):564-72. doi: 10.1016/j.coi.2011.08.001. Epub 2011 Aug 22. Curr Opin Immunol. 2011. PMID: 21865020 Review. - IκB kinase ε (IKKε): a therapeutic target in inflammation and cancer.
Verhelst K, Verstrepen L, Carpentier I, Beyaert R. Verhelst K, et al. Biochem Pharmacol. 2013 Apr 1;85(7):873-80. doi: 10.1016/j.bcp.2013.01.007. Epub 2013 Jan 17. Biochem Pharmacol. 2013. PMID: 23333767 Free PMC article. Review.
Cited by
- MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors.
Castanier C, Zemirli N, Portier A, Garcin D, Bidère N, Vazquez A, Arnoult D. Castanier C, et al. BMC Biol. 2012 May 24;10:44. doi: 10.1186/1741-7007-10-44. BMC Biol. 2012. PMID: 22626058 Free PMC article. - MST4 negatively regulates type I interferons production via targeting MAVS-mediated pathway.
Liu W, Ma Z, Wu Y, Yuan C, Zhang Y, Liang Z, Yang Y, Zhang W, Jiao P. Liu W, et al. Cell Commun Signal. 2022 Jul 12;20(1):103. doi: 10.1186/s12964-022-00922-3. Cell Commun Signal. 2022. PMID: 35820905 Free PMC article. - Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41.
Su C, Zheng C. Su C, et al. J Virol. 2017 Feb 28;91(6):e02414-16. doi: 10.1128/JVI.02414-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077645 Free PMC article. - Regulation of cellular innate antiviral signaling by ubiquitin modification.
Lin D, Zhong B. Lin D, et al. Acta Biochim Biophys Sin (Shanghai). 2015 Mar;47(3):149-55. doi: 10.1093/abbs/gmu133. Epub 2015 Feb 3. Acta Biochim Biophys Sin (Shanghai). 2015. PMID: 25651846 Free PMC article. Review. - An Integrated View of Deubiquitinating Enzymes Involved in Type I Interferon Signaling, Host Defense and Antiviral Activities.
Qian G, Zhu L, Li G, Liu Y, Zhang Z, Pan J, Lv H. Qian G, et al. Front Immunol. 2021 Oct 11;12:742542. doi: 10.3389/fimmu.2021.742542. eCollection 2021. Front Immunol. 2021. PMID: 34707613 Free PMC article. Review.
References
- Arguello, M. D., and J. Hiscott. 2007. Ub surprised: viral ovarian tumor domain proteases remove ubiquitin and ISG15 conjugates. Cell Host Microbe 2367-369. - PubMed
- Bibeau-Poirier, A., S. P. Gravel, J. F. Clement, S. Rolland, G. Rodier, P. Coulombe, J. Hiscott, N. Grandvaux, S. Meloche, and M. J. Servant. 2006. Involvement of the IκB kinase (IKK)-related kinases Tank-binding kinase 1/IKKi and Cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 1775059-5067. - PubMed
- Bibeau-Poirier, A., and M. J. Servant. 2008. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine 43359-367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous