A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB - PubMed (original) (raw)
A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB
G J Bean et al. Biochemistry. 2009.
Abstract
S-(3,4-Dichlorobenzyl)isothiourea (A22) disrupts the actin cytoskeleton of bacteria, causing defects of morphology and chromosome segregation. Previous studies have suggested that the actin homologue MreB itself is the target of A22, but there has been no direct observation of A22 binding to MreB and no mechanistic explanation of its mode of action. We show that A22 binds MreB with at least micromolar affinity in its nucleotide-binding pocket in a manner that is sterically incompatible with simultaneous ATP binding. A22 negatively affects both the time course and extent of MreB polymerization in vitro in the presence of ATP. A22 prevents assembly of MreB into long, rigid polymers, as determined by both fluorescence microscopy and sedimentation assays. A22 increases the critical concentration of ATP-bound MreB assembly from 500 nM to approximately 2000 nM. We therefore conclude that A22 is a competitive inhibitor of ATP binding to MreB. A22-bound MreB is capable of polymerization, but with assembly properties that more closely resemble those of the ADP-bound state. Because the cellular concentration of MreB is in the low micromolar range, this mechanism explains the ability of A22 to largely disassemble the actin cytoskeleton in bacterial cells. It also represents a novel mode of action for a cytoskeletal drug and the first biochemical characterization of the interaction between a small molecule inhibitor of the bacterial cytoskeleton and its target.
Figures
Figure 1. A22-MreB interaction observed by NMR
5 mM A22 was incubated with varying concentrations of MreB in 10 mM Tris-HCl, pH 8.0 at 10°C. Normalized integrated peak areas collected for the three aromatic protons in A22 were plotted versus protein concentration and fitted to a one-site binding model. The Kd was found to be 1.32 ± 0.14 µM (R2 = 0.89).
Figure 2. A22 docking in the MreB nucleotide cleft
(A) Docking site of A22 in the nucleotide-free form of MreB. (B) Details of the binding site of A22 in MreB with residues within 2.5 Å of A22 shown with their sidechains. ATP is shown in its crystal position, illustrating the clashes that would occur between its beta and gamma phosphates and A22.
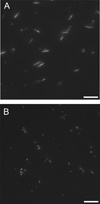
Figure 3. A22-mediated inhibition of MreB assembly visualized by fluorescence microscopy
1 µM MreB (20% Alexa-488 labeled) was polymerized in 10 mM imidazole, pH 7.0, 1 mM MgCl2, 1 mM EGTA, 20 mM KCl and 200 µM ATP at 20° C for 1 hr in the absence (A) or presence (B) of 300 µM A22 and imaged directly by epifluorescence microscopy. Scale bar = 10 µm.
Figure 4. Effects of A22 on MreB polymerization timecourse
5 µM MreB was polymerized in 10 mM imidazole, pH 7.0, 20 mM KCl, 1 mM MgCl2 and 200 µM ATP at 20° C in the presence of varying concentrations of A22. MreB polymerization timecourse was followed by 400 nm right angle light scattering. A22 concentrations are as specified in the inset.
Figure 5. Effect of A22 on MreB critical concentration
Varying concentrations of MreB were polymerized at 4° C overnight in 10 mM imidazole, pH 7.0, 1 mM MgCl2, 1 mM EGTA, 20 mM KCl and 200 µM ATP in the absence (A) or presence (B) of 300 µM A22. Samples were equilibrated to 20° C for 1 h and the 400 nm light scattering intensity was measured. Linear fits to the data yielded critical concentrations of 500 nM (A) and 2000 nM (B), respectively.
Figure 5. Effect of A22 on MreB critical concentration
Varying concentrations of MreB were polymerized at 4° C overnight in 10 mM imidazole, pH 7.0, 1 mM MgCl2, 1 mM EGTA, 20 mM KCl and 200 µM ATP in the absence (A) or presence (B) of 300 µM A22. Samples were equilibrated to 20° C for 1 h and the 400 nm light scattering intensity was measured. Linear fits to the data yielded critical concentrations of 500 nM (A) and 2000 nM (B), respectively.
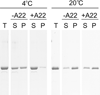
Figure 6. Effects of A22 on sedimentation of MreB
5 µM MreB was polymerized for 1 h at 4° C or 20° C in 10 mM imidazole, pH 7.0, 1 mM MgCl2, 1 mM EGTA, 20 mM KCl and 200 µM ATP in the presence or absence of 100 µM A22. Samples were centrifuged at 4° C or 20° C for 30 min at 100000g. Equivalent volumes of total (T), supernatant (S) and pellet (P) samples are shown by SDS-PAGE and Coomassie blue.
Similar articles
- Polymerization properties of the Thermotoga maritima actin MreB: roles of temperature, nucleotides, and ions.
Bean GJ, Amann KJ. Bean GJ, et al. Biochemistry. 2008 Jan 15;47(2):826-35. doi: 10.1021/bi701538e. Epub 2007 Dec 21. Biochemistry. 2008. PMID: 18095710 Free PMC article. - Exploring the A22-Bacterial Actin MreB Interaction through Molecular Dynamics Simulations.
Awuni Y, Jiang S, Robinson RC, Mu Y. Awuni Y, et al. J Phys Chem B. 2016 Sep 22;120(37):9867-74. doi: 10.1021/acs.jpcb.6b05199. Epub 2016 Sep 14. J Phys Chem B. 2016. PMID: 27600765 - The assembly of MreB, a prokaryotic homolog of actin.
Esue O, Cordero M, Wirtz D, Tseng Y. Esue O, et al. J Biol Chem. 2005 Jan 28;280(4):2628-35. doi: 10.1074/jbc.M410298200. Epub 2004 Nov 16. J Biol Chem. 2005. PMID: 15548516 - The actin-like MreB proteins in Bacillus subtilis: a new turn.
Chastanet A, Carballido-Lopez R. Chastanet A, et al. Front Biosci (Schol Ed). 2012 Jun 1;4(4):1582-606. doi: 10.2741/s354. Front Biosci (Schol Ed). 2012. PMID: 22652894 Review. - Bacterial DNA segregation by the actin-like MreB protein.
Kruse T, Gerdes K. Kruse T, et al. Trends Cell Biol. 2005 Jul;15(7):343-5. doi: 10.1016/j.tcb.2005.05.002. Trends Cell Biol. 2005. PMID: 15922599 Review.
Cited by
- Concerning Increase in Antimicrobial Resistance in Shiga Toxin-Producing Escherichia coli Isolated from Young Animals during 1980-2016.
Chirila F, Tabaran A, Fit N, Nadas G, Mihaiu M, Tabaran F, Cătoi C, Reget OL, Dan SD. Chirila F, et al. Microbes Environ. 2017 Sep 27;32(3):252-259. doi: 10.1264/jsme2.ME17023. Epub 2017 Sep 12. Microbes Environ. 2017. PMID: 28904263 Free PMC article. - The molecular origins of chiral growth in walled cells.
Huang KC, Ehrhardt DW, Shaevitz JW. Huang KC, et al. Curr Opin Microbiol. 2012 Dec;15(6):707-14. doi: 10.1016/j.mib.2012.11.002. Epub 2012 Nov 26. Curr Opin Microbiol. 2012. PMID: 23194654 Free PMC article. Review. - Omnipose: a high-precision morphology-independent solution for bacterial cell segmentation.
Cutler KJ, Stringer C, Lo TW, Rappez L, Stroustrup N, Brook Peterson S, Wiggins PA, Mougous JD. Cutler KJ, et al. Nat Methods. 2022 Nov;19(11):1438-1448. doi: 10.1038/s41592-022-01639-4. Epub 2022 Oct 17. Nat Methods. 2022. PMID: 36253643 Free PMC article. - ParB proteins can bypass DNA-bound roadblocks via dimer-dimer recruitment.
Tišma M, Panoukidou M, Antar H, Soh YM, Barth R, Pradhan B, Barth A, van der Torre J, Michieletto D, Gruber S, Dekker C. Tišma M, et al. Sci Adv. 2022 Jul;8(26):eabn3299. doi: 10.1126/sciadv.abn3299. Epub 2022 Jun 29. Sci Adv. 2022. PMID: 35767606 Free PMC article. - Virtual Screening Guided Design, Synthesis and Bioactivity Study of Benzisoselenazolones (BISAs) on Inhibition of c-Met and Its Downstream Signalling Pathways.
Zhang S, Song Q, Wang X, Wei Z, Yu R, Wang X, Jiang T. Zhang S, et al. Int J Mol Sci. 2019 May 20;20(10):2489. doi: 10.3390/ijms20102489. Int J Mol Sci. 2019. PMID: 31137515 Free PMC article.
References
- Jones LJF, Carballido-Lopez R, Errington J. Control of Cell Shape in Bacteria: Helical, Actin-like Filaments in Bacillus subtilis. Cell. 2001;104:913–922. - PubMed
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. - PubMed
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The Bacterial Cytoskeleton: An Intermediate Filament-Like Function in Cell Shape. Cell. 2003;115:705–713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM067246/GM/NIGMS NIH HHS/United States
- P41 GM066326/GM/NIGMS NIH HHS/United States
- P41RR02301/RR/NCRR NIH HHS/United States
- P41GM66326/GM/NIGMS NIH HHS/United States
- R01 GM067246-05/GM/NIGMS NIH HHS/United States
- S10 RR002781/RR/NCRR NIH HHS/United States
- S10 RR008438/RR/NCRR NIH HHS/United States
- R01 GM067246/GM/NIGMS NIH HHS/United States
- P41 RR002301/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases