The role of histone deacetylases (HDACs) in human cancer - PubMed (original) (raw)
Review
The role of histone deacetylases (HDACs) in human cancer
Santiago Ropero et al. Mol Oncol. 2007 Jun.
Abstract
The balance of histone acetylation and deacetylation is an epigenetic layer with a critical role in the regulation of gene expression. Histone acetylation induced by histone acetyl transferases (HATs) is associated with gene transcription, while histone hypoacetylation induced by histone deacetylase (HDAC) activity is associated with gene silencing. Altered expression and mutations of genes that encode HDACs have been linked to tumor development since they both induce the aberrant transcription of key genes regulating important cellular functions such as cell proliferation, cell-cycle regulation and apoptosis. Thus, HDACs are among the most promising therapeutic targets for cancer treatment, and they have inspired researchers to study and develop HDAC inhibitors.
Figures
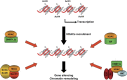
Figure 1
Different ways by which HDACs are recruited to gene promoters. An array of nucleosomes is shown. Histone octamers are represented by circles and the DNA is shown in red. Histone deacetylation induced by recruitment of HDAC to gene promoters by different factors including, DNA methyl transferases (DNMT), the methyl binding protein MeCP2, Estrogen receptor (ER) and transcription factors (E2F, Rb).
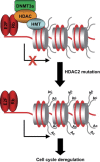
Figure 2
A model showing a possible effect of HDAC2 mutation in cancer development. class I HDACs are involved in gene transcription‐repression mediated by retinoblastoma protein. The lost of HDAC2 function could induce the hyperacetylation and reexpression of genes regulated by retinoblastoma protein Rb, and with crucial functions in cell cycle regulation.
Similar articles
- HDACs and HDAC Inhibitors in Cancer Development and Therapy.
Li Y, Seto E. Li Y, et al. Cold Spring Harb Perspect Med. 2016 Oct 3;6(10):a026831. doi: 10.1101/cshperspect.a026831. Cold Spring Harb Perspect Med. 2016. PMID: 27599530 Free PMC article. Review. - Protein deacetylases: enzymes with functional diversity as novel therapeutic targets.
Yoshida M, Shimazu T, Matsuyama A. Yoshida M, et al. Prog Cell Cycle Res. 2003;5:269-78. Prog Cell Cycle Res. 2003. PMID: 14593721 Review. - Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy.
Ramaiah MJ, Tangutur AD, Manyam RR. Ramaiah MJ, et al. Life Sci. 2021 Jul 15;277:119504. doi: 10.1016/j.lfs.2021.119504. Epub 2021 Apr 16. Life Sci. 2021. PMID: 33872660 Review. - Focus on acetylation: the role of histone deacetylase inhibitors in cancer therapy and beyond.
Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Konstantinopoulos PA, et al. Expert Opin Investig Drugs. 2007 May;16(5):569-71. doi: 10.1517/13543784.16.5.569. Expert Opin Investig Drugs. 2007. PMID: 17461732 Review. - Histone deacetylases and cancer: causes and therapies.
Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Marks P, et al. Nat Rev Cancer. 2001 Dec;1(3):194-202. doi: 10.1038/35106079. Nat Rev Cancer. 2001. PMID: 11902574 Review.
Cited by
- Modular Development of Enzyme-Activatable Proteolysis Targeting Chimeras for Selective Protein Degradation and Cancer Targeting.
Chen Y, Zhang L, Fang L, Chen C, Zhang D, Peng T. Chen Y, et al. JACS Au. 2024 May 16;4(7):2564-2577. doi: 10.1021/jacsau.4c00298. eCollection 2024 Jul 22. JACS Au. 2024. PMID: 39055140 Free PMC article. - Design, synthesis and biological evaluation of 2,4-pyrimidinediamine derivatives as ALK and HDACs dual inhibitors for the treatment of ALK addicted cancer.
Guo D, Yu Y, Long B, Deng P, Ran D, Han L, Zheng J, Gan Z. Guo D, et al. J Enzyme Inhib Med Chem. 2022 Dec;37(1):2512-2529. doi: 10.1080/14756366.2022.2121822. J Enzyme Inhib Med Chem. 2022. PMID: 36100230 Free PMC article. - Anti-multiple myeloma potential of resynthesized belinostat derivatives: an experimental study on cytotoxic activity, drug combination, and docking studies.
Nguyen HP, Tran Q, Nguyen CQ, Hoa TP, Duy Binh T, Nhu Thao H, Hue BTB, Tuan NT, Le Dang Q, Quoc Chau Thanh N, Van Ky N, Pham MQ, Yang SG. Nguyen HP, et al. RSC Adv. 2022 Aug 10;12(34):22108-22118. doi: 10.1039/d2ra01969h. eCollection 2022 Aug 4. RSC Adv. 2022. PMID: 36043105 Free PMC article. - HDAC1 Expression in Invasive Ductal Carcinoma of the Breast and Its Value as a Good Prognostic Factor.
Eom M, Oh SS, Lkhagvadorj S, Han A, Park KH. Eom M, et al. Korean J Pathol. 2012 Aug;46(4):311-7. doi: 10.4132/KoreanJPathol.2012.46.4.311. Epub 2012 Aug 23. Korean J Pathol. 2012. PMID: 23110022 Free PMC article. - The Dynamics of Interactions Among Immune and Glioblastoma Cells.
Eder K, Kalman B. Eder K, et al. Neuromolecular Med. 2015 Dec;17(4):335-52. doi: 10.1007/s12017-015-8362-x. Epub 2015 Jul 30. Neuromolecular Med. 2015. PMID: 26224516 Review.
References
- Bouras, T. , Fu, M. , Sauve, A.A. , Wang, F. , Quong, A.A. , Perkins, N.D. , Hay, R.T. , Gu, W. , Pestell, R.G. , 2005. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem.. 280, 10264–10276. - PubMed
- Bradbury, C.A. , Khanim, F.L. , Hayden, R. , Bunce, C.M. , White, D.A. , Drayson, M.T. , Craddock, C. , Turner, B.M. , 2005. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 19, 1751–1759. - PubMed
- Brehm, A. , Miska, E.A. , McCance, D.J. , Reid, J.L. , Bannister, A.J. , Kouzarides, T. , 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 391, 597–601. - PubMed
- Chen, W.Y. , Wang, D.H. , Yen, R.C. , Luo, J. , Gu, W. , Baylin, S.B. , 2005. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 123, 437–448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases