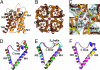Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance - PubMed (original) (raw)
Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance
Joseph D Ho et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Aquaporin (AQP) 4 is the predominant water channel in the mammalian brain, abundantly expressed in the blood-brain and brain-cerebrospinal fluid interfaces of glial cells. Its function in cerebral water balance has implications in neuropathological disorders, including brain edema, stroke, and head injuries. The 1.8-A crystal structure reveals the molecular basis for the water selectivity of the channel. Unlike the case in the structures of water-selective AQPs AqpZ and AQP1, the asparagines of the 2 Asn-Pro-Ala motifs do not hydrogen bond to the same water molecule; instead, they bond to 2 different water molecules in the center of the channel. Molecular dynamics simulations were performed to ask how this observation bears on the proposed mechanisms for how AQPs remain totally insulating to any proton conductance while maintaining a single file of hydrogen bonded water molecules throughout the channel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
General features. (A and B) Monomer and tetramer views of hAQP4 in diagram representation. Brown and orange colors represent the N- and C-terminal pseudo 2-fold related portions. Water molecules are represented as red spheres, and glycerol molecules are shown as green sticks. (A) The side view of the monomer. Helices are labeled M1 to M8. (B) The tetramer viewed from the extracellular side down the crystallographic 4-fold symmetry axis. (C) The network of water molecules found at the intracellular side of the central pore. The central pore is at the crystallographic 4-fold symmetry axis, and is formed by the tetramer. The 2Fo-Fc density of the water molecules is shown in black, contoured at 1.2 σ. The backbone amides of Ser-188 and Gly-189 are colored yellow in diagram representation. Phe-195 is shown as brown stick and cyan surface. (D–F) Diagram representation of the C loop of all of the AQP X-ray structures solved to date. (D) E. coli GlpF (brown), archeal AqpM (magenta), spinach AQP SoPIP2;1 (blue), and PfAQP (green). (E) Rat AQP4 (yellow), human AQP4 (black), human AQP5 (red), E. coli AqpZ (cyan), bovine AQP0 (green), and bovine AQP1 (purple). (F) Comparison of the 310 helix of rat AQP4 (yellow) with the 2-turn helix of AqpM (magenta) and spinach AQP (blue). All structural renderings were made with PyMOL (
).
Fig. 2.
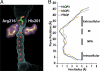
The conducting pore. The trace of the pore inner surface is shown in cyan. The selectivity filter residues, Arg-216 and His-201, are shown as sticks with surfaces in purple. The glycerol molecule is shown as green stick, and the water molecules in the channel are shown as red spheres. (B) Plot of the channel radius versus position along the pore for human AQP4, bovine AQP1 (bAQP1), and the P. falciparum AQP (PfAQP). Regions of the channel are labeled as extracellular vestibule, the selectivity filter (SF), the NPA motif, and the intracellular vestibule. The pore inner surface and its dimension are calculated using Hole 2.0 (51).
Fig. 3.
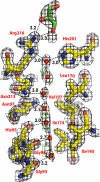
Electron density. Residues that form the wall of the pore are shown in sticks. Water molecules are shown as red spheres. The glycerol molecule is shown in green stick. The 2Fo-Fc density is shown in black, contoured at 1.2 σ. Positive _F_o − _F_c density is shown in green, contoured at 3 σ. There is no negative _F_o − _F_c density.
Fig. 4.
Comparison of the hydrogen bond network of the selectivity filter arginine of hAQP4, bAQP1, and GlpF. Protein C-alpha is shown in diagram representation. Residues of the selectivity filter and glycerol molecules are shown as sticks. Water molecules are shown as red spheres.
Fig. 5.
The NPA motifs. (A) Schematic representation of the hydrogen bonding network through the channels of hAQP4 and bAQP1. The distances are between heavy-atom to heavy-atom. (B) Stick representation of the NPA motifs. Distances that are too long to be a hydrogen bond are colored in red. (C) Plot of the MD simulations of hAQP4 from 4 different experiments. Details are described in Discussion.
Similar articles
- Mechanism of aquaporin-4's fast and highly selective water conduction and proton exclusion.
Tani K, Mitsuma T, Hiroaki Y, Kamegawa A, Nishikawa K, Tanimura Y, Fujiyoshi Y. Tani K, et al. J Mol Biol. 2009 Jun 19;389(4):694-706. doi: 10.1016/j.jmb.2009.04.049. Epub 2009 May 3. J Mol Biol. 2009. PMID: 19406128 - Structural Determinants of Oligomerization of the Aquaporin-4 Channel.
Kitchen P, Conner MT, Bill RM, Conner AC. Kitchen P, et al. J Biol Chem. 2016 Mar 25;291(13):6858-71. doi: 10.1074/jbc.M115.694729. Epub 2016 Jan 19. J Biol Chem. 2016. PMID: 26786101 Free PMC article. - Water transport in human aquaporin-4: molecular dynamics (MD) simulations.
Cui Y, Bastien DA. Cui Y, et al. Biochem Biophys Res Commun. 2011 Sep 9;412(4):654-9. doi: 10.1016/j.bbrc.2011.08.019. Epub 2011 Aug 12. Biochem Biophys Res Commun. 2011. PMID: 21856282 Free PMC article. - Selectivity and conductance among the glycerol and water conducting aquaporin family of channels.
Stroud RM, Savage D, Miercke LJ, Lee JK, Khademi S, Harries W. Stroud RM, et al. FEBS Lett. 2003 Nov 27;555(1):79-84. doi: 10.1016/s0014-5793(03)01195-5. FEBS Lett. 2003. PMID: 14630323 Review. - Astrocyte Aquaporin Dynamics in Health and Disease.
Potokar M, Jorgačevski J, Zorec R. Potokar M, et al. Int J Mol Sci. 2016 Jul 13;17(7):1121. doi: 10.3390/ijms17071121. Int J Mol Sci. 2016. PMID: 27420057 Free PMC article. Review.
Cited by
- Arterial pulsations drive oscillatory flow of CSF but not directional pumping.
Kedarasetti RT, Drew PJ, Costanzo F. Kedarasetti RT, et al. Sci Rep. 2020 Jun 22;10(1):10102. doi: 10.1038/s41598-020-66887-w. Sci Rep. 2020. PMID: 32572120 Free PMC article. - Reversible, temperature-dependent supramolecular assembly of aquaporin-4 orthogonal arrays in live cell membranes.
Crane JM, Verkman AS. Crane JM, et al. Biophys J. 2009 Dec 2;97(11):3010-8. doi: 10.1016/j.bpj.2009.09.017. Biophys J. 2009. PMID: 19948131 Free PMC article. - Immunodominant T cell determinants of aquaporin-4, the autoantigen associated with neuromyelitis optica.
Nelson PA, Khodadoust M, Prodhomme T, Spencer C, Patarroyo JC, Varrin-Doyer M, Ho JD, Stroud RM, Zamvil SS. Nelson PA, et al. PLoS One. 2010 Nov 30;5(11):e15050. doi: 10.1371/journal.pone.0015050. PLoS One. 2010. PMID: 21151500 Free PMC article. - Identification of a point mutation impairing the binding between aquaporin-4 and neuromyelitis optica autoantibodies.
Pisani F, Mola MG, Simone L, Rosito S, Alberga D, Mangiatordi GF, Lattanzi G, Nicolotti O, Frigeri A, Svelto M, Nicchia GP. Pisani F, et al. J Biol Chem. 2014 Oct 31;289(44):30578-30589. doi: 10.1074/jbc.M114.582221. Epub 2014 Sep 19. J Biol Chem. 2014. PMID: 25239624 Free PMC article. - Tetra detector analysis of membrane proteins.
Miercke LJW, Robbins RA, Stroud RM. Miercke LJW, et al. Curr Protoc Protein Sci. 2014 Aug 1;77:29.10.1-29.10.30. doi: 10.1002/0471140864.ps2910s77. Curr Protoc Protein Sci. 2014. PMID: 25081744 Free PMC article. Review.
References
- Rojek A, Praetorius J, Frokiaer J, Nielsen S, Fenton RA. A current view of the mammalian aquaglyceroporins. Annu Rev Physiol. 2008;70:301–327. - PubMed
- King LS, Kozono D, Agre P. From structure to disease: The evolving tale of aquaporin biology. Nat Rev Mol Cell Biol. 2004;5:687–698. - PubMed
- Badaut J, Brunet JF, Regli L. Aquaporins in the brain: From aqueduct to “multi-duct.”. Metab Brain Dis. 2007;22:251–263. - PubMed
- Manley GT, et al. Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat Med. 2000;6:159–163. - PubMed
- Badaut J, Lasbennes F, Magistretti PJ, Regli L. Aquaporins in Brain: Distribution, Physiology, and Pathophysiology. J Cereb Blood Flow Metab. 2002;22:367–378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM024485/GM/NIGMS NIH HHS/United States
- R01 GM024485/GM/NIGMS NIH HHS/United States
- GM24485/GM/NIGMS NIH HHS/United States
- GM074929/GM/NIGMS NIH HHS/United States
- P50 GM073210/GM/NIGMS NIH HHS/United States
- U54 GM074929/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases