Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies - PubMed (original) (raw)
. 2009 May 1;137(3):509-21.
doi: 10.1016/j.cell.2009.04.027. Epub 2009 Apr 23.
Vasily V Vagin, Soohyun Lee, Jia Xu, Shengmei Ma, Hualin Xi, Hervé Seitz, Michael D Horwich, Monika Syrzycka, Barry M Honda, Ellen L W Kittler, Maria L Zapp, Carla Klattenhoff, Nadine Schulz, William E Theurkauf, Zhiping Weng, Phillip D Zamore
Affiliations
- PMID: 19395009
- PMCID: PMC2768572
- DOI: 10.1016/j.cell.2009.04.027
Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies
Chengjian Li et al. Cell. 2009.
Abstract
Piwi-interacting RNAs (piRNAs) silence transposons in animal germ cells. piRNAs are thought to derive from long transcripts spanning transposon-rich genomic loci and to direct an autoamplification loop in which an antisense piRNA, bound to Aubergine or Piwi protein, triggers production of a sense piRNA bound to the PIWI protein Argonaute3 (Ago3). In turn, the new piRNA is envisioned to produce a second antisense piRNA. Here, we describe strong loss-of-function mutations in ago3, allowing a direct genetic test of this model. We find that Ago3 acts to amplify piRNA pools and to enforce on them an antisense bias, increasing the number of piRNAs that can act to silence transposons. We also detect a second, Ago3-independent piRNA pathway centered on Piwi. Transposons targeted by this second pathway often reside in the flamenco locus, which is expressed in somatic ovarian follicle cells, suggesting a role for piRNAs beyond the germline.
Figures
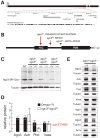
Figure 1
ago3 mutants. (A) The ago3 gene resides in pericentromeric heterochromatin on the left arm of chromosome 3. (B) ago3 mutant alleles were identified by TILLING. (C) Full-length Ago3 protein was not detected in trans-heterozygous ago3 ovaries, but was readily detected in heterozygotes and wild-type. (D) Protein levels in wild-type and ago3 ovaries, relative to ago3/TM6B (red line). The average ± standard deviation for at least three independent biological samples is shown. (E) Representative data for Aub, Piwi, Vasa, Argonaute1 (Ago1), Argonaute2 (Ago2), and Armitage (Armi) in ago3 ovaries. Tubulin served as a loading control.
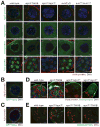
Figure 2
Consequences of loss of Ago3 in the Drosophila germ line. (A) Mutually interdependent incorporation of Ago3 and Aub, into nuage in ovaries. Images correspond to single confocal sections (63× magnification); scale bar: 5 μm (Ago3, Aub, and Vasa panels) or 10 μm (Piwi). (B) and (C) Vasa localization within nuage is only partially congruent with that of Ago3 and Aub. Images correspond to single confocal sections of stage 4/5 egg chambers (63× lens, 4× zoom). Scale bars: 10 μm. (D) In 5−7 day old male adults, Vasa-expressing germ-line stem cells (GSC), which normally surround the somatic hub cells (hub), were not detected in ago3 testes. Since Vasa expression increases in ago3, the two images from ago3 mutants were acquired using reduced gain relative to wild-type and heterozygous testes. Scale bar: 10 μm (first three images) or 20 μm (fourth). (E) Stellate silencing in testes requires Ago3. Scale bars: 20 μm.
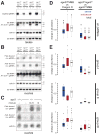
Figure 3
Aub- and Piwi-bound piRNAs disappear without Ago3. (A) Accumulation of Su(Ste) and AT-chX-1 piRNAs in testes required Ago3. (B) Accumulation of roo antisense piRNAs, as well as the AT-chX-1 piRNA, in ovaries required Ago3. (C) Accumulation of Piwi- and Aub-bound roo antisense piRNAs in the ovary was rescued in ago3 mutants by a single-copy transgene expressing Ago3 in the germ line (G) or in both the germ line and the soma (G+S). (D) Box plots illustrating the change in abundance of piRNAs, analyzed by transposon family, in ago3/TM6B versus Oregon R (left panel) and ago3 versus ago3/TM6B (right panel) ovaries. (E) Box plots illustrating the change in abundance for piRNAs for each of 95 transposon families, separated by group.
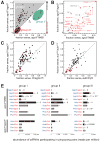
Figure 4
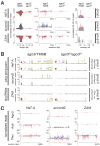
Three distinct groups of piRNAs. (A) The sense fraction of the piRNAs was compared for each transposon family between ago3 and ago3/TM6B ovaries. (B) An enlargement of the lower left corner of (A). Transposons labeled in red are present in the flamenco locus. Here and in (C) and (D), group III transposon families not present in flamenco are denoted with a red-filled black circle. (C) The sense fraction of piRNAs was compared between ago3 and Oregon R ovaries. (D) The sense fraction of the piRNAs was compared between aub and aub/CyO ovaries. (E) The normalized abundance of ping-pong pairs detected for all nine (ago3/TM6B) or four (ago3) possible PIWI protein pairings for the piRNA species uniquely associated with a single PIWI protein. Black bars, Bonferroni-corrected _p_-value < 0.005; gray bars, _p_-value > 0.005. _p_-values < 10-30 are reported as p ∼ 0. Values are average ± two standard deviations.
Figure 5
Paradigmatic examples of each transposon group. (A) piRNA length distribution, abundance relative to consensus position, and ping-pong pair abundance. Blue, sense piRNAs; red, antisense. (B) Sequence-excess logos generated by subtracting the background from the relative frequency of each nucleotide at each position of the 23–29 nt RNAs in each sample. Only the nucleotide positions where foreground was significantly higher than background are shown (Bonferroni-corrected _p_-values < 0.001). Gray horizontal bars indicate the maximum possible value for each position. (C) Normalized and calibrated abundance of piRNAs uniquely associated with Ago3, Aub, or Piwi in ago3/TM6B or ago3 ovaries.
Figure 6
Loss of Ago3 increases expression of some group I and group III, but not group II, transposons. (A) Expression of group I, group II, and group III transposon families in ago3 ovaries, relative to wild-type (w1118) ovaries, was assayed using whole-genome tiling microarrays. White bars: expression of the transposon family in both ago3/TM6B and ago3 ovaries was less than that of the 50th percentile for expression of all mRNAs in wild-type, suggesting that expression change cannot be reliably quantified. Black bars: transposon families with expression greater than this threshold in one or both genotypes. Significant (false-discovery rate (FDR) < 0.02) and non-significant data (FDR > 0.02) are separated. (B) Quantitative RT-PCR was used to assess transposon expression, relative to Actin, for ago3 ovaries (black bars) and ovaries expressing one copy of a UAS-Ago3 transgene driven by _nanos_-Gal4 (gray bars) or _actin5c_-Gal4 (white bars), relative to ago3/TM6B ovaries.
Figure 7
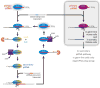
A model for piRNA biogenesis. The Aub- and Ago3-dependent piRNA amplification cycle is envisioned to operate only in the germ line, whereas a Piwi-dependent, Aub- and Ago3-independent pathway is shown for somatic cells. In the germ line, Piwi can also partner with Ago3 to amplify piRNAs.
Similar articles
- Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms.
Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. Wang W, et al. Mol Cell. 2015 Sep 3;59(5):819-30. doi: 10.1016/j.molcel.2015.08.007. Mol Cell. 2015. PMID: 26340424 Free PMC article. - Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production.
Han BW, Wang W, Li C, Weng Z, Zamore PD. Han BW, et al. Science. 2015 May 15;348(6236):817-21. doi: 10.1126/science.aaa1264. Science. 2015. PMID: 25977554 Free PMC article. - Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis.
Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Nagao A, et al. RNA. 2010 Dec;16(12):2503-15. doi: 10.1261/rna.2270710. Epub 2010 Oct 27. RNA. 2010. PMID: 20980675 Free PMC article. - piRNA-mediated silencing in Drosophila germlines.
Siomi MC, Miyoshi T, Siomi H. Siomi MC, et al. Semin Cell Dev Biol. 2010 Sep;21(7):754-9. doi: 10.1016/j.semcdb.2010.01.011. Epub 2010 Jan 18. Semin Cell Dev Biol. 2010. PMID: 20080197 Review. - piRNA pathway and the potential processing site, the nuage, in the Drosophila germline.
Pek JW, Patil VS, Kai T. Pek JW, et al. Dev Growth Differ. 2012 Jan;54(1):66-77. doi: 10.1111/j.1440-169x.2011.01316.x. Dev Growth Differ. 2012. PMID: 23741748 Review.
Cited by
- UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery.
Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE. Zhang F, et al. Cell. 2012 Nov 9;151(4):871-884. doi: 10.1016/j.cell.2012.09.040. Cell. 2012. PMID: 23141543 Free PMC article. - PIWI-interacting RNAs: from generation to transgenerational epigenetics.
Luteijn MJ, Ketting RF. Luteijn MJ, et al. Nat Rev Genet. 2013 Aug;14(8):523-34. doi: 10.1038/nrg3495. Epub 2013 Jun 25. Nat Rev Genet. 2013. PMID: 23797853 Review. - Germ granules in spermatogenesis of Drosophila: Evidences of contribution to the piRNA silencing.
Kibanov MV, Gvozdev VA, Olenina LV. Kibanov MV, et al. Commun Integr Biol. 2012 Mar 1;5(2):130-3. doi: 10.4161/cib.18741. Commun Integr Biol. 2012. PMID: 22808315 Free PMC article. - Reexamining the P-Element Invasion of Drosophila melanogaster Through the Lens of piRNA Silencing.
Kelleher ES. Kelleher ES. Genetics. 2016 Aug;203(4):1513-31. doi: 10.1534/genetics.115.184119. Genetics. 2016. PMID: 27516614 Free PMC article. Review. - Emerging roles and potential application of PIWI-interacting RNA in urological tumors.
Zhang J, Zhang W, Liu Y, Pi M, Jiang Y, Ainiwaer A, Mao S, Chen H, Ran Y, Sun S, Li W, Yao X, Chang Z, Yan Y. Zhang J, et al. Front Endocrinol (Lausanne). 2023 Jan 17;13:1054216. doi: 10.3389/fendo.2022.1054216. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36733811 Free PMC article. Review.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. - PubMed
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM65236/GM/NIGMS NIH HHS/United States
- R01 GM062862/GM/NIGMS NIH HHS/United States
- GM050898/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM065236/GM/NIGMS NIH HHS/United States
- GM62862/GM/NIGMS NIH HHS/United States
- HD049116/HD/NICHD NIH HHS/United States
- R01 HD049116/HD/NICHD NIH HHS/United States
- R37 GM062862/GM/NIGMS NIH HHS/United States
- R01 GM050898/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials