Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate - PubMed (original) (raw)
doi: 10.1038/ng.370. Epub 2009 Apr 26.
Jennifer Tran, Anuradha Gopalan, Zhenbang Chen, Safa Shaikh, Arkaitz Carracedo, Andrea Alimonti, Caterina Nardella, Shohreh Varmeh, Peter T Scardino, Carlos Cordon-Cardo, William Gerald, Pier Paolo Pandolfi
Affiliations
- PMID: 19396168
- PMCID: PMC2835150
- DOI: 10.1038/ng.370
Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate
Brett S Carver et al. Nat Genet. 2009 May.
Erratum in
- Author Correction: Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate.
Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. Carver BS, et al. Nat Genet. 2020 Sep;52(9):984. doi: 10.1038/s41588-020-0688-0. Nat Genet. 2020. PMID: 32820258
Abstract
Chromosomal translocations involving the ERG locus are frequent events in human prostate cancer pathogenesis; however, the biological role of aberrant ERG expression is controversial. Here we show that aberrant expression of ERG is a progression event in prostate tumorigenesis. We find that prostate cancer specimens containing the TMPRSS2-ERG rearrangement are significantly enriched for loss of the tumor suppressor PTEN. In concordance with these findings, transgenic overexpression of ERG in mouse prostate tissue promotes marked acceleration and progression of high-grade prostatic intraepithelial neoplasia (HGPIN) to prostatic adenocarcinoma in a Pten heterozygous background. In vitro overexpression of ERG promotes cell migration, a property necessary for tumorigenesis, without affecting proliferation. ADAMTS1 and CXCR4, two candidate genes strongly associated with cell migration, were upregulated in the presence of ERG overexpression. Thus, ERG has a distinct role in prostate cancer progression and cooperates with PTEN haploinsufficiency to promote progression of HGPIN to invasive adenocarcinoma.
Figures
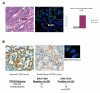
Figure 1. Genetic and molecular alterations ERG and PTEN are frequent and concomitant events in human prostate cancer
FISH break apart probes to 3′ (red) and 5′ (green) ends of both ERG and TMPRSS2 were used. Microscopy (200X), showing hematoxylin and eosin staining of HGPIN (arrowhead) and prostate carcinoma juxtaposed, high grade PIN (arrowhead) with deletion of the 3′ TMPRSS2 region, and prostatic adenocarcinoma (insert) with deletion of the 3′ TMPRSS2 region, as seen by loss of the red signal in at least one of the red-green pairs (A). Bar graph shows the proportion of HGPIN cases by ERG status (A). Tumor tissue microarray demonstrating prostate cancer with normal PTEN staining and prostate cancer with weak/absent PTEN staining (B). FISH staining from consecutive sections of the same specimen demonstrating the ERG genetic rearrangement is shown. The table reports the frequency of PTEN status by ERG rearrangement status (B).
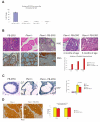
Figure 2. Prostate specific over-expression of ERG cooperates with Pten haploinsufficiency to promote prostate tumorigenesis
Locally advanced mouse (_Pten_−/− _p53_−/−) prostate cancer demonstrated increased expression of murine Erg at the transcript level (normalized to Hprt) compared to controls (A). A total of 12 Pten+/−, 12 ERG transgenic mice, and 12 Pten+/− ERG mice were phenotypically characterized. High (200X) power histologic demonstration of prostatic adenocarcinoma in Pten+/− ERG mice compared to controls in mice 6 months of age (B). Immunohistochemistry for smooth muscle actin is performed demonstrating invasion (B). Phenotypic characterization of Pten+/− ERG mice at 2 and 6 months of age (200X). The incidence of prostatic adenocarcinoma is reported for transgenic mice at mice 2, 4, and 6 months of age (B). A bar graph is shown demonstrating the percentage of mice at each time point with the histologic finding of prostatic adenocarcinoma. Immunohistochemistry for Ki67 staining in pre-neoplastic prostate glands (mice 4 months of age) demonstrated no increase in proliferation for transgenic ERG mice while Pten haploinsufficiency provides a proliferative advantage (C). A bar graph is shown demonstrating the percentage of ki67 positive staining cells per gland with mean and standard deviation reported for 3 representative prostate glands with p-value calculated based on Chi-square statistic (C). Immunohistochemistry of Pten revealed no differences in staining for pre-neoplastic prostate glands at 4 months of age and prostate in vivo transcript levels for Pten and AKT were similar for both genotypes (D).
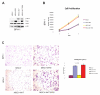
Figure 3. ERG regulates cell migration
Western blot analysis determined expression of Flag-ERG construct and activation of P-AKT in stable BPH-1 cell lines infected with MSCV, MSCV-ERG, MSCV-AKT (AKT-1), and MSCV-AKT MSCV-ERG (A). While over-expression of a constitutively active AKT (AKT-1) promoted cell growth, no proliferative advantage was observed with ERG over-expression (B). A line graph is utilized to demonstrate cell proliferation with the mean and standard deviation represented from 3 experiments (B). A striking increase was observed in cell migration with over-expression of ERG compared to vector control in BPH-1 cells which was not augmented with over-expression of a constitutively active AKT (AKT-1) (C). Bar graphs demonstrate the fold change in the number of migrated cells per high powered field normalized to vector control, and the mean and standard deviation from 3 experiments were calculated with p-value calculated based on total number of cells migrating (C).
Figure 4. ERG directly regulates CXCR4 and ADAMTS1
Putative ETS binding sites were identified in the promoter regions of both human CXCR4 and ADAMTS1 genes (A). ChIP assays were performed demonstrating direct ERG binding to the promoter regions for both CXCR4 and ADAMTS1 (A). Repeat ChIP experiment for CXCR4 in PC3 cells over-expressing ERG (B). CXCR4 siRNA knock-down demonstrates a significant reduction in cell migration in PC3 cells over-expressing ERG (C). Further evaluation revealed that the mRNA expression of murine Cxcr4 was up-regulated in the prostate specimens of ERG transgenic mice (D) and Pten loss models of prostate tumorigenesis with up-regulation of ERG compared to controls (E). The bar graphs demonstrate the fold change in mRNA level following normalization to Hprt and our lowest value, and the mean and standard deviations from 3 experiments are shown.
Comment in
- Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis.
King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL. King JC, et al. Nat Genet. 2009 May;41(5):524-6. doi: 10.1038/ng.371. Epub 2009 Apr 26. Nat Genet. 2009. PMID: 19396167 Free PMC article. - TMPRSS2-ERG and PTEN loss in prostate cancer.
Squire JA. Squire JA. Nat Genet. 2009 May;41(5):509-10. doi: 10.1038/ng0509-509. Nat Genet. 2009. PMID: 19399032 - Words of wisdom. Re: Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate.
Bubendorf L. Bubendorf L. Eur Urol. 2009 Nov;56(5):882-3. doi: 10.1016/j.eururo.2009.08.007. Eur Urol. 2009. PMID: 20965031 No abstract available.
Similar articles
- Molecular evidence that invasive adenocarcinoma can mimic prostatic intraepithelial neoplasia (PIN) and intraductal carcinoma through retrograde glandular colonization.
Haffner MC, Weier C, Xu MM, Vaghasia A, Gürel B, Gümüşkaya B, Esopi DM, Fedor H, Tan HL, Kulac I, Hicks J, Isaacs WB, Lotan TL, Nelson WG, Yegnasubramanian S, De Marzo AM. Haffner MC, et al. J Pathol. 2016 Jan;238(1):31-41. doi: 10.1002/path.4628. Epub 2015 Oct 14. J Pathol. 2016. PMID: 26331372 Free PMC article. - βIII-tubulin overexpression is an independent predictor of prostate cancer progression tightly linked to ERG fusion status and PTEN deletion.
Tsourlakis MC, Weigand P, Grupp K, Kluth M, Steurer S, Schlomm T, Graefen M, Huland H, Salomon G, Steuber T, Wilczak W, Sirma H, Simon R, Sauter G, Minner S, Quaas A. Tsourlakis MC, et al. Am J Pathol. 2014 Mar;184(3):609-17. doi: 10.1016/j.ajpath.2013.11.007. Epub 2013 Dec 28. Am J Pathol. 2014. PMID: 24378408 - Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion.
Gumuskaya B, Gurel B, Fedor H, Tan HL, Weier CA, Hicks JL, Haffner MC, Lotan TL, De Marzo AM. Gumuskaya B, et al. Prostate Cancer Prostatic Dis. 2013 Jun;16(2):209-15. doi: 10.1038/pcan.2013.8. Epub 2013 Apr 2. Prostate Cancer Prostatic Dis. 2013. PMID: 23545904 Free PMC article. - ERG protein expression as a biomarker of prostate cancer.
Falzarano SM, Magi-Galluzzi C. Falzarano SM, et al. Biomark Med. 2013 Dec;7(6):851-65. doi: 10.2217/bmm.13.105. Biomark Med. 2013. PMID: 24266818 Review. - Clinical applications of novel ERG immunohistochemistry in prostate cancer diagnosis and management.
Shah RB. Shah RB. Adv Anat Pathol. 2013 Mar;20(2):117-24. doi: 10.1097/PAP.0b013e3182862ac5. Adv Anat Pathol. 2013. PMID: 23399797 Review.
Cited by
- Nanomedicines in diagnosis and treatment of prostate cancers: an updated review.
Wang J, Zhang X, Xing J, Gao L, Lu H. Wang J, et al. Front Bioeng Biotechnol. 2024 Aug 21;12:1444201. doi: 10.3389/fbioe.2024.1444201. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39318666 Free PMC article. Review. - JUN mediates the senescence associated secretory phenotype and immune cell recruitment to prevent prostate cancer progression.
Redmer T, Raigel M, Sternberg C, Ziegler R, Probst C, Lindner D, Aufinger A, Limberger T, Trachtova K, Kodajova P, Högler S, Schlederer M, Stoiber S, Oberhuber M, Bolis M, Neubauer HA, Miranda S, Tomberger M, Harbusch NS, Garces de Los Fayos Alonso I, Sternberg F, Moriggl R, Theurillat JP, Tichy B, Bystry V, Persson JL, Mathas S, Aberger F, Strobl B, Pospisilova S, Merkel O, Egger G, Lagger S, Kenner L. Redmer T, et al. Mol Cancer. 2024 May 29;23(1):114. doi: 10.1186/s12943-024-02022-x. Mol Cancer. 2024. PMID: 38811984 Free PMC article. - Identification of genes that promote PI3K pathway activation and prostate tumour formation.
Francis JC, Capper A, Rust AG, Ferro K, Ning J, Yuan W, de Bono J, Pettitt SJ, Swain A. Francis JC, et al. Oncogene. 2024 Jun;43(24):1824-1835. doi: 10.1038/s41388-024-03028-x. Epub 2024 Apr 23. Oncogene. 2024. PMID: 38654106 Free PMC article. - ERG activates a stem-like proliferation-differentiation program in prostate epithelial cells with mixed basal-luminal identity.
Feng W, Ladewig E, Salsabeel N, Zhao H, Lee YS, Gopalan A, Lange M, Luo H, Kang W, Fan N, Rosiek E, de Stanchina E, Chen Y, Carver BS, Leslie CS, Sawyers CL. Feng W, et al. bioRxiv [Preprint]. 2024 Apr 6:2023.05.15.540839. doi: 10.1101/2023.05.15.540839. bioRxiv. 2024. PMID: 38585869 Free PMC article. Preprint. - Retinoic acid receptor activation reprograms senescence response and enhances anti-tumor activity of natural killer cells.
Colucci M, Zumerle S, Bressan S, Gianfanti F, Troiani M, Valdata A, D'Ambrosio M, Pasquini E, Varesi A, Cogo F, Mosole S, Dongilli C, Desbats MA, Contu L, Revankdar A, Chen J, Kalathur M, Perciato ML, Basilotta R, Endre L, Schauer S, Othman A, Guccini I, Saponaro M, Maraccani L, Bancaro N, Lai P, Liu L, Pernigoni N, Mele F, Merler S, Trotman LC, Guarda G, Calì B, Montopoli M, Alimonti A. Colucci M, et al. Cancer Cell. 2024 Apr 8;42(4):646-661.e9. doi: 10.1016/j.ccell.2024.02.004. Epub 2024 Feb 29. Cancer Cell. 2024. PMID: 38428412 Free PMC article.
References
- Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. - PubMed
- Tu JJ, et al. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod. Pathol. 2007;20:921–928. - PubMed
- Perner S, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am. J. Surg. Pathol. 2007;31:882–888. - PubMed
- Hermans KG, et al. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. - PubMed
- Helgeson BE, et al. Characterization of TMPRSS2:ETV4 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA082328/CA/NCI NIH HHS/United States
- R01 MD004038/MD/NIMHD NIH HHS/United States
- P50 CA092629-09/CA/NCI NIH HHS/United States
- P50-CA92629/CA/NCI NIH HHS/United States
- P50 CA092629/CA/NCI NIH HHS/United States
- R01-CA84292/CA/NCI NIH HHS/United States
- R01-CA82328/CA/NCI NIH HHS/United States
- R01 CA082328-12/CA/NCI NIH HHS/United States
- U01 CA084292-10/CA/NCI NIH HHS/United States
- U01 CA084292/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous