Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont - PubMed (original) (raw)
Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont
Eric C Martens et al. J Biol Chem. 2009.
Abstract
Bacteria in the distal human gut have evolved diverse abilities to metabolize complex glycans, including the capacity to degrade these compounds as nutrients and to assemble their component sugars into new polymers such as extracellular capsules. The human gut bacterium Bacteroides thetaiotaomicron is well endowed with the ability to metabolize both host- and diet-derived glycans. Its genome contains 88 different polysaccharide utilization loci (PULs) for complex glycan catabolism and eight different gene clusters for capsular polysaccharide biosynthesis. Here, we investigate one of the prominent mechanisms by which this gut symbiont regulates many PULs involved in host mucin O-glycan degradation; namely, transcriptional regulation via the concerted interactions of cell-envelope-localized TonB-dependent transporters, extra-cytoplasmic function sigma factors and anti-sigma factors, which participate together in a regulatory pathway termed trans-envelope signaling. Unexpectedly, we found that several different trans-envelope signaling switches involved in PUL-mediated O-glycan degradation also modulate capsular polysaccharide synthesis. A novel regulatory pathway, which is dependent on expression of O-glycan-targeting outer membrane proteins, governs this coordinated regulation of glycan catabolism and capsule synthesis. This latter finding provides a new link in the dynamic interplay between complex glycan metabolism, microbial physiology, and host responses that occurs during colonization of the gut.
Figures
FIGURE 1.
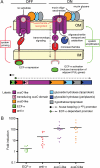
Positive feedback model of PUL regulation. A, schematic of a hypothetical Sus-like system regulated by trans-envelope signaling and the PUL at which it is encoded (note that gene number and enzyme content is variable between different PULs). The left half of the Fig. illustrates the sequence of events leading to system priming in the “off” signaling state: (i) initial expression of ECF-σ (green) from a housekeeping σAB promoter (dashed arrow with a black circular base) results in a small amount of PUL gene expression via the ECF-σ-dependent transcript; (ii) simultaneous production of anti-σ (red), which physically interacts with both the SusC-like transporter (purple) and the cytoplasmic ECF-σ, causes the system to be held in the default off state unless glycan substrates are present. The right half illustrates events leading to amplified expression (“on” state) of the Sus-like system: (i) a cognate substrate (e.g. mucin _O_-glycan) is bound and then transported through the transducing SusC-like transporter; (ii) substrate transport initiates an allosteric signal that is transmitted from the SusC-like protein to the anti-σ factor via periplasmic contacts and then to the ECF-σ factor via cytoplasmic contacts; (iii) propagation of the trans-envelope signal results in ECF-σ activation and increased expression of the ECF-σ-dependent promoter(s) (dashed arrow with a green circular base) located upstream of nearby genes encoding structural and enzymatic components of the Sus-like system. B, comparison of induction levels for 12 trans-envelope signaling systems that respond to host glycans. The -fold induction (plotted on the y axis) relative to MM-glucose is shown for genes encoding ECF-σ factors (green data points), anti-σ factors (red data points), transducing SusC-like proteins (purple data points), and SusD-like proteins (orange data points) from 12 different PULs that are regulated by trans-envelope signaling. Data points represent individual genes, and horizontal black bars represent average -fold induction for each gene class in GeneChip expression experiments. The y axis is shown on log10 scale to allow visualization of differences in ECF-σ expression.
FIGURE 2.
Protein interactions between trans-envelope signaling components. A, yeast two-hybrid assays of interactions between periplasmic trans-envelope signaling components (SusC-like protein N-terminal transducing domains and anti-σ C-terminal domains). B, yeast two-hybrid assays of interactions between cytoplasmic trans-envelope signaling components (ECF-σ factors and anti-σ N-terminal domains). Each grid represents a matrix of 25 different interactions tested for each signaling node. Panels on the left show growth of all possible interacting pairs under non-selective conditions that do not require protein interactions (Leu-, Trp-). Panels on the right show replicate growths of the same interacting partners shown on the left but under selective conditions (Leu-, Trp-, Ade-, His-); colony growth under this condition indicates protein interaction in the assay. Cognate interactions are highlighted with red circles, and non-cognate interactions are highlighted with blue circles. Gene names at the top and left of columns/rows indicate the source genes tested. The precise coding sequences used for yeast two-hybrid partner fusions are illustrated in supplemental Fig.
S2
. Colony growth was photographed after 2 days of growth at 30 °C.
FIGURE 3.
Molecular genetic analysis of trans-envelope signaling. A, schematic of the BT1032–53 PUL, which is induced in response to mucin _O_-glycans. Key genes that were subjected to mutational analysis are indicated with locus tag numbers, and all genes are labeled according to the key provided in Fig. 1; white boxes in this schematic represent genes with annotated functions other than those indicated in Fig. 1. B, a histogram showing the responses of 22 genes contained within the BT1032–53 PUL to different growth and/or bacterial genetic backgrounds. For wild-type B. thetaiotaomicron grown in a preparation of neutral _O_-glycans (14), most genes in this PUL show increased expression relative to a MM-glucose reference (yellow bars). Individual disruptions of genes encoding either the signal transducing SusC-like protein (BT1042, purple bars) or the ECF-σ associated with this system (BT1053, green bars) result in nearly complete loss of PUL activation during growth in the neutral _O_-glycan mixture. Conversely, de-repression of the trans-envelope signaling switch via anti-σ mutation (red bars) results in PUL expression during growth on glucose as a sole carbon source, which does not ordinarily cause induction of these genes. Values are based on normalized GeneChip expression data. Bars indicate -fold changes in expression relative to duplicate reference datasets derived from 5-ml cultures grown in MM-glucose. All bars represent the average response of each gene in two replicate datasets.
FIGURE 4.
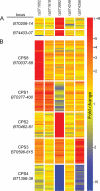
Other loci regulated by ECF-σ factors. A heatmap illustrating the transcriptional responses of genes in two PULs, BT0206–14 and BT4403–07 (A), and five B. thetaiotaomicron CPS synthesis loci (B) to mutation of five individual anti-σ factors and consequent de-repression of their corresponding ECF-σ-dependent regulons. Responses of individual genes within the locus tag range noted for each cluster are shown and are based on -fold change values determined from GeneChip datasets. Three biological replicates were performed for each anti-σ mutant and -fold changes quantified by referencing each gene to the average expression values observed in three datasets obtained from wild-type B. thetaiotaomicron grown in MM-glucose. -Fold change values are calibrated to the color bar shown at the right. Note that each of the PULs shown in panel A exhibits a specific response to disruption of anti-σBT3992, which activates ECF-σBT3993 but not other regulators. Also, four different CPS loci (CPS8, CPS1, CPS2, and CPS3) are activated differently in the various anti-σ mutants. One locus, CPS4, exhibits transcriptional repression under two conditions. Three remaining B. thetaiotaomicron VPI-5482 capsule loci (CPS5–7), which did not respond to any of the conditions tested, are not shown.
FIGURE 5.
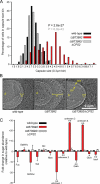
Capsule production by the Ω_BT3992_ mutant. A, histogram plot of capsule sizes determined by India ink staining of the isogenic wild-type (black bars), Ω_BT3992_ (red bars), and the Ω_BT3992/Δ_CPS2 double mutant (gray bars) strains. Capsule sizes from a total of 150 individual cells were measured for each strain, 50 from each of three different experiments. p values are provided above the Ω_BT3992_ distribution and are color-coded based on the dataset (wild-type or double mutant) to which the Ω_BT3992_ distribution was compared. Representative images of India ink-stained encapsulated cells are provided in supplemental Fig.
S5
. B, quick-freeze, deep-etch scanning electron micrographs of wild-type and Ω_BT3992_ and Ω_BT3992/Δ_CPS2 double mutant cells, illustrating their capsule morphology. The capsule appears thicker in the Ω_BT3992_ mutant but of similar density as the other two strains. The width of each capsule (c) is indicated by a yellow line. The bacterial cell (b), inner membrane (im), and outer membrane (om) are labeled for reference. Magnification is 50,000×. C, analysis of monosaccharides present in wild-type and Ω_BT3992_ and Ω_BT3992/Δ_CPS2 double mutant capsules. A total of 13 different sugars were differentiated by HPAEC-PAD; nine of these eluted at the same time as known monosaccharide standards (see labels above histogram bars), whereas four compounds (labeled as unknown 1–4) did not correspond to any known standards used. Note the abundance of two uniquely represented and unknown sugars (unknowns 3 and 4) in the Ω_BT3992_ capsule. Total extracellular polysaccharides were extracted from cells grown in MM-glucose, and three biological replicates were performed for each strain. Values shown represent the average -fold increase of each sugar relative to the average present in wild-type cells, and error bars represent the S.D. Instances where sugar abundance in the Ω_BT3992_ mutant differed significantly from wild-type are indicated with asterisks (*, p ≤ 0.05; **, p ≤ 0.01 by Student's t test). Histogram bars are arranged along the x axis based on their elution points under the HPAEC-PAD conditions used (negative charge increases toward the right side of the graph). Representative HPAEC-PAD traces are provided in supplemental Fig.
S6
. Fuc, fucose; GalA, galacturonic acid; GlcA, glucuronic acid; IdoA, iduronic acid.
FIGURE 6.
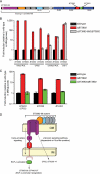
Different regulatory pathways mediate CPS locus and PUL expression. A, a schematic of the BT3983–94 PUL. ORFs are color-coded according to the legend in Fig. 1, and predicted glycoside hydrolase (GH) activities are provided based on CAZy family designations. The white ORF in the middle of the locus is BT3989, a hypothetical gene that does not exhibit induction with the adjacent genes and may not be involved in PUL function. B, quantitative reverse transcriptase-PCR analysis of gene expression from three loci in wild-type (black bars) and Ω_BT3992_ (red bars) and Ω_BT3992_/Δ_BT3983–88_ (gray bars) strain backgrounds. Two genes, BT3983 and BT3984, encoding outer membrane protein components of the BT3983–94 Sus-like system, exhibit strong induction when anti-σBT3992 is disrupted. Note that these two transcripts are absent and, therefore, are not detectable in the Ω_BT3992_/Δ_BT3983–88_ double mutant. Similarly, two genes (BT0463 and BT0482) positioned near the beginning and end of CPS2, show induction in the Ω_BT3992_ strain. However, this induction is reduced to wild-type levels in the Ω_BT3992_/Δ_BT3983–88_ double mutant, indicating that expression of the BT3983–88 Sus-like components is required for CPS activation. A probe gene (BT0206) from one PUL (BT0206–14) that is unlinked to BT3992 is also dependent on expression of the BT3983–88 for its activation. Conversely, induction of a transcript (BT4404) from another unlinked PUL (BT4403–07) in both mutant strains indicates that the induction of unlinked genes is not always dependent on the presence of outer membrane components. Similarly, regulation of BT3992 is unaffected by loss of the Sus-like proteins. C, expression of genes in the ECF-σBT3993 regulon in isogenic wild-type (black bars) and Ω_BT3992_ (red bars) and Ω_BT3992_/Δ_BT3993_ (green bars) strains. Loss of ECF-σBT3993 eliminates activation of all genes in the regulon. Most notably, expression of BT4404, a component of the ECF-σ-linked BT4403–07 PUL, is reduced to wild-type levels. Thus, the cross-talk between these loci must occur at the level of ECF-σBT3993 activation of the BT4404 promoter and not at other, non-cognate signaling nodes, such as anti-σBT3992 activation of ECF-σBT4403, which would still be intact in the Ω_BT3992_/Δ_BT3993_ strain. D, a model summarizing the results of data presented in panels B and C. Note that the unknown signaling pathway that controls CPS2 and BT0206–14 expression is also dependent on the trans-envelope signaling pathway, which is required for expression of the BT3983–88 Sus-like system. IM, inner membrane; OM, outer membrane.
References
- Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A. ( 2008) Nat. Rev. Microbiol. 6, 121– 131 - PubMed
- Peterson D. A., McNulty N. P., Guruge J. L., Gordon J. I. ( 2007) Cell Host Microbe 2, 328– 339 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK030292/DK/NIDDK NIH HHS/United States
- F32 AI073060/AI/NIAID NIH HHS/United States
- T32 HD07409/HD/NICHD NIH HHS/United States
- DK30292/DK/NIDDK NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases