An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein - PubMed (original) (raw)
An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein
Xin-Jian He et al. Cell. 2009.
Abstract
DNA methylation is a conserved epigenetic mark in plants and mammals. In Arabidopsis, DNA methylation can be triggered by small interfering RNAs (siRNAs) through an RNA-directed DNA methylation (RdDM) pathway. Here, we report the identification of an RdDM effector, KTF1. Loss-of-function mutations in KTF1 reduce DNA methylation and release the silencing of RdDM target loci without abolishing the siRNA triggers. KTF1 has similarity to the transcription elongation factor SPT5 and contains a C-terminal extension rich in GW/WG repeats. KTF1 colocalizes with ARGONAUTE 4 (AGO4) in punctate nuclear foci and binds AGO4 and RNA transcripts. Our results suggest KTF1 as an adaptor protein that binds scaffold transcripts generated by Pol V and recruits AGO4 and AGO4-bound siRNAs to form an RdDM effector complex. The dual interaction of an effector protein with AGO and small RNA target transcripts may be a general feature of RNA-silencing effector complexes.
Figures
Figure 1. Transcriptional gene silencing of RD29A-LUC is suppressed by the rdm3-1 and rdm3-2 mutations
(A) Effect of rdm3-1 and rdm3-2 on luminescence and kanamycin-resistance phenotypes in the ros1 background. Plants were grown on MS plates and subjected to luminescence imaging after cold treatment (4°C, 24 h). The plants were also grown on MS plates with kanamycin (50 μg/ml) and photographed after 10 days. (B) Northern blot analysis of RNA levels of endogenous RD29A, RD29A-LUC, and 35S-NPTII in wild type, ros1, and ros1rdm3-1. The constitutively expressed 18S rRNA was used as an RNA loading control while COR15A was used as a cold treatment control.
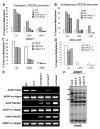
Figure 2. The rdm3 mutations reduce DNA methylation at RdDM target loci
The percentage of cytosine methylation was determined by bisulfite sequencing at transgenic (A) and endogenous (B) RD29A promoters, AtSN1 (C) and MEA-ISR (D). The percentage of cytosine methylation on CG, CHG, and CHH sites is shown. H represents A, T, or C. (E) The rdm3-1 mutation suppressed DNA methylation in AtSN1, AtGP1, and AtMU1. After the indicated genomic DNA was digested with the methylation-sensitive restriction enzyme HaeIII, it was used for amplification of AtSN1. After the genomic DNA was digested with the methylated DNA-specific restriction enzyme McrBC, it was used for amplification of AtGP1 and AtMU1. The amplifications of non-digested genomic DNA were used as controls. (F) The rdm3-3 mutation reduced AtMU1 methylation at CHH sites. Genomic DNA from the indicated genotypes was digested with HaeIII, followed by Southern blot analysis. The three undigested bands (arrows) that are present in the Col-0 wild type were mostly digested in rdm3-3, nrpd1-3 and nrpe1-11.
Figure 3. Effect of the rdm3 mutations on RNA and siRNA levels from the RdDM target loci
(A) The rdm3 mutations increase the RNA expression levels of AtSN1, AtGP1, and AtMU1. Semi-quantitative RT-PCR was used to detect the transcript levels of AtSN1 (interval A, see diagram in panel C), AtGP1, and AtMU1 in the indicated genotypes. TUB8 was amplified as an internal control. (B) Small RNA blot analysis of 24-nt siRNAs, 21-nt ta-siRNAs, and microRNAs in the various genotypes. The positions of size markers (21 nt and 24 nt) are indicated. The ethidium bromide-stained small RNA gel is shown as a loading control. (C) Strand-specific RT-PCR analysis of IGN5, IGN6, AtSN1 and solo LTR transcripts in the Col-0 wild type, nrpe1-11 and rdm3-3. Actin PCR products and total RNA resolved by agarose gel electrophoresis serve as loading controls. Reactions without reverse transcriptase (no RT) were performed to control for background DNA contamination. The positions of the different AtSN1 intervals tested by RT-PCR are indicated in the diagram on the left. (D) RT-PCR analysis of RD29A promoter transcript. TUB8 and ethidium bromide-stained gel are shown as controls. (E) RT-PCR detection of RD29A promoter transcript in KTF1 immunoprecipitates. The background signal from TUB8 was used as an internal control, which indicated no difference between the RNA amounts from ros1 and ros1rdm3-1. No AB, controls without using anti-KTF1 antibody.
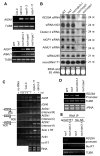
Figure 4. The WG/GW repeats in KTF1 C-terminal domain interact with AGO4
(A) KTF1, NRPE1, HsGW182 and proteins are shown schematically. All three proteins are characterized by the reiterated WG/GW repeats-containing domains (in yellow). The red stripes represent each of the WG/GW repeat. WG-1 and WG-2 represent two highly conserved WG repeat regions in KTF1. (B) Diagram of the bacterially expressed NRPE1-CTD and truncated KTF1 proteins. (C) The purified proteins were subjected to SDS-PAGE and gels were stained with Coomassie. Arrows point to the proteins of interest. (D) Western blot analysis showing that the GST-fused truncated KTF1 and NRPE1-CTD interact with Myc-AGO4 from plant extracts. Ten percent of the input was used in the “Input” lane. (E) Anti-Myc antibody-conjugated beads captured truncated KTF1 proteins and NRPE1-CTD from a mixture of the proteins with extract from Myc-AGO4 plants. Arrows point to the proteins of interest. (F) and (G) Western blot analysis showing coimmunoprecipitation of KTF1 and Myc-AGO4. Ler wild-type plants without the Myc-AGO4 transgene were used as controls. No AB, control precipitation without using antibodies.
Figure 5. Sub-nuclear localization of KTF1 in interphase Arabidopsis nuclei
A. Detection of KTF1 (in red) in wild type (WT) and rdm3-1 mutant nuclei by immunofluorescence using anti-KTF1. B. Simultaneous localization of KTF1 and AGO4 or NRPE1. KTF1 (red) was localized using its specific antibody in cells expressing cMyc- and Flag-tagged AGO4 and NRPE1 (in green), respectively. The bright yellow signals due to the overlap of red and green channels in merged images indicate colocalization of two labeled proteins. In all panels DNA was stained with DAPI (blue). Size bar corresponds to 5μm.
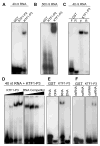
Figure 6. The KTF1 C-terminal domain binds RNAs
(A) KTF1-P3 but not KTF1-P2 binds to a 40-nt RNA (corresponding to the RD29A promoter) in electrophoretic mobility shift assays. (B) KTF1-P3 but not KTF1-P2 binds to a 500-nt RNA corresponding to the RD29A promoter. (C) KTF1-P3 binds to both the forward (F) and reverse (R) strands of the 40-nt RNA. (D) Protein concentration-dependence of the RNA-binding and competition by unlabeled RNA. The protein-RNA complex increased when an increasing amount of KTF1-P3 protein (0.2, 0.4, 0.8, 1.6 μg) was added to the binding reaction. The protein-RNA complex decreased when an increasing amount of unlabeled 40-nt RNA (1x, 5x, 25x, 125x of labeled RNA) was added to the binding reaction. (E) KTF1-P3 binds to the 40-nt RNA but does not bind to DNA of the same sequence. (F) KTF1-P3 binds to the single-stranded but not double-stranded 40-nt RNA.
Similar articles
- NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses.
Raja P, Jackel JN, Li S, Heard IM, Bisaro DM. Raja P, et al. J Virol. 2014 Mar;88(5):2611-22. doi: 10.1128/JVI.02305-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352449 Free PMC article. - Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation.
Duan CG, Zhang H, Tang K, Zhu X, Qian W, Hou YJ, Wang B, Lang Z, Zhao Y, Wang X, Wang P, Zhou J, Liang G, Liu N, Wang C, Zhu JK. Duan CG, et al. EMBO J. 2015 Mar 4;34(5):581-92. doi: 10.15252/embj.201489453. Epub 2014 Dec 19. EMBO J. 2015. PMID: 25527293 Free PMC article. - RNA-directed DNA methylation in plants: Where to start?
Zhang H, He X, Zhu JK. Zhang H, et al. RNA Biol. 2013 Oct;10(10):1593-6. doi: 10.4161/rna.26312. RNA Biol. 2013. PMID: 25003825 Free PMC article. Review. - Molecular mechanisms of the RNA polymerases in plant RNA-directed DNA methylation.
Xie G, Du X, Hu H, Du J. Xie G, et al. Trends Biochem Sci. 2024 Mar;49(3):247-256. doi: 10.1016/j.tibs.2023.11.005. Epub 2023 Dec 9. Trends Biochem Sci. 2024. PMID: 38072749 Review.
Cited by
- Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review. - Nuclear pyruvate dehydrogenase complex regulates histone acetylation and transcriptional regulation in the ethylene response.
Shao Z, Bian L, Ahmadi SK, Daniel TJ, Belmonte MA, Burns JG, Kotla P, Bi Y, Shen Z, Xu SL, Wang ZY, Briggs SP, Qiao H. Shao Z, et al. Sci Adv. 2024 Jul 26;10(30):eado2825. doi: 10.1126/sciadv.ado2825. Epub 2024 Jul 26. Sci Adv. 2024. PMID: 39058774 Free PMC article. - A conserved Pol II elongator SPT6L mediates Pol V transcription to regulate RNA-directed DNA methylation in Arabidopsis.
Liu Y, Shu J, Zhang Z, Ding N, Liu J, Liu J, Cui Y, Wang C, Chen C. Liu Y, et al. Nat Commun. 2024 May 25;15(1):4460. doi: 10.1038/s41467-024-48940-8. Nat Commun. 2024. PMID: 38796517 Free PMC article. - Citrus psorosis virus 24K protein inhibits the processing of miRNA precursors by interacting with components of the biogenesis machinery.
Marmisolle FE, Borniego MB, Cambiagno DA, Gonzalo L, García ML, Manavella PA, Hernández C, Reyes CA. Marmisolle FE, et al. Microbiol Spectr. 2024 Jul 2;12(7):e0351323. doi: 10.1128/spectrum.03513-23. Epub 2024 May 24. Microbiol Spectr. 2024. PMID: 38785434 Free PMC article. - DNA-dependent RNA polymerases in plants.
Yang DL, Huang K, Deng D, Zeng Y, Wang Z, Zhang Y. Yang DL, et al. Plant Cell. 2023 Sep 27;35(10):3641-3661. doi: 10.1093/plcell/koad195. Plant Cell. 2023. PMID: 37453082 Free PMC article.
References
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. - PubMed
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. - PubMed
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM059138/GM/NIGMS NIH HHS/United States
- R01 GM070795/GM/NIGMS NIH HHS/United States
- R01 GM070795-06/GM/NIGMS NIH HHS/United States
- GM077590/GM/NIGMS NIH HHS/United States
- R01 GM059138/GM/NIGMS NIH HHS/United States
- R01 GM059138-11/GM/NIGMS NIH HHS/United States
- R01GM070795/GM/NIGMS NIH HHS/United States
- R01 GM077590/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases