The basal initiation machinery: beyond the general transcription factors - PubMed (original) (raw)
Review
The basal initiation machinery: beyond the general transcription factors
Timothy W Sikorski et al. Curr Opin Cell Biol. 2009 Jun.
Abstract
In vitro experiments led to a simple model in which basal transcription factors sequentially assembled with RNA Polymerase II to generate a preinitiation complex (PIC). Emerging evidence indicates that PIC composition is not universal, but promoter-dependent. Active promoters are occupied by a mixed population of complexes, including regulatory factors such as NC2, Mot1, Mediator, and TFIIS. Recent studies are expanding our understanding of the roles of these factors, demonstrating that their functions are both broader and more context dependent than previously realized.
Figures
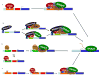
Figure. Many Paths to the PIC
The factors and assembly pathways used to form transcriptionally competent preinitiation complexes can be promoter dependent [3,73]. 1) TBP assembling onto promoter regions via TFIID leads to recruitment of the other basal initiation factors, as outlined in the stepwise assembly pathway [1]. In S. cerevisiae, this pathway is most often utilized at TATA-less genes. At some mammalian promoters, histone H3K4 trimethylation helps to recruit the TFIID complex [5]. 2) Mediator bridges interactions between activators and the basal initiation machinery, and can stimulate basal transcription as well. At some promoters Mediator can recruit TFIIH and TFIIE independently of RNApII [45]. 3) TBP can also be brought to promoters by the SAGA complex. In S. cerevisiae, this pathway is most utilized at TATA containing promoters. The Mot1 and NC2 complexes can repress this pathway by actively removing TBP from the TATA element [73]. 4) Mot1 and NC2 can also have a positive role in transcription by removing nonproductive TBP complexes from DNA, thereby allowing functional PICs to form [60,65,77].
Similar articles
- Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae.
Geisberg JV, Moqtaderi Z, Kuras L, Struhl K. Geisberg JV, et al. Mol Cell Biol. 2002 Dec;22(23):8122-34. doi: 10.1128/MCB.22.23.8122-8134.2002. Mol Cell Biol. 2002. PMID: 12417716 Free PMC article. - The general transcription machinery and general cofactors.
Thomas MC, Chiang CM. Thomas MC, et al. Crit Rev Biochem Mol Biol. 2006 May-Jun;41(3):105-78. doi: 10.1080/10409230600648736. Crit Rev Biochem Mol Biol. 2006. PMID: 16858867 Review. - Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo.
Geisberg JV, Holstege FC, Young RA, Struhl K. Geisberg JV, et al. Mol Cell Biol. 2001 Apr;21(8):2736-42. doi: 10.1128/MCB.21.8.2736-2742.2001. Mol Cell Biol. 2001. PMID: 11283253 Free PMC article. - The capping enzyme facilitates promoter escape and assembly of a follow-on preinitiation complex for reinitiation.
Fujiwara R, Damodaren N, Wilusz JE, Murakami K. Fujiwara R, et al. Proc Natl Acad Sci U S A. 2019 Nov 5;116(45):22573-22582. doi: 10.1073/pnas.1905449116. Epub 2019 Oct 7. Proc Natl Acad Sci U S A. 2019. PMID: 31591205 Free PMC article. - Interplay between the transcription preinitiation complex and the +1 nucleosome.
Chen X, Xu Y. Chen X, et al. Trends Biochem Sci. 2024 Feb;49(2):145-155. doi: 10.1016/j.tibs.2023.12.001. Epub 2024 Jan 13. Trends Biochem Sci. 2024. PMID: 38218671 Review.
Cited by
- A Conserved Nuclear Cyclophilin Is Required for Both RNA Polymerase II Elongation and Co-transcriptional Splicing in Caenorhabditis elegans.
Ahn JH, Rechsteiner A, Strome S, Kelly WG. Ahn JH, et al. PLoS Genet. 2016 Aug 19;12(8):e1006227. doi: 10.1371/journal.pgen.1006227. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27541139 Free PMC article. - A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting.
Tantale K, Mueller F, Kozulic-Pirher A, Lesne A, Victor JM, Robert MC, Capozi S, Chouaib R, Bäcker V, Mateos-Langerak J, Darzacq X, Zimmer C, Basyuk E, Bertrand E. Tantale K, et al. Nat Commun. 2016 Jul 27;7:12248. doi: 10.1038/ncomms12248. Nat Commun. 2016. PMID: 27461529 Free PMC article. - Mechanisms and consequences of alternative polyadenylation.
Di Giammartino DC, Nishida K, Manley JL. Di Giammartino DC, et al. Mol Cell. 2011 Sep 16;43(6):853-66. doi: 10.1016/j.molcel.2011.08.017. Mol Cell. 2011. PMID: 21925375 Free PMC article. Review. - Transcription inhibition as a therapeutic target for cancer.
Stellrecht CM, Chen LS. Stellrecht CM, et al. Cancers (Basel). 2011 Nov 23;3(4):4170-90. doi: 10.3390/cancers3044170. Cancers (Basel). 2011. PMID: 24213132 Free PMC article. - The role of cotranscriptional histone methylations.
Buratowski S, Kim T. Buratowski S, et al. Cold Spring Harb Symp Quant Biol. 2010;75:95-102. doi: 10.1101/sqb.2010.75.036. Epub 2011 Mar 29. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21447819 Free PMC article.
References
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. - PubMed
- Reese JC. Basal transcription factors. Curr Opin Genet Dev. 2003;13:114–118. - PubMed
- Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem. 2007;282:14685–14689. This review discusses the large variety of promoter types found throughout the genome and provide models for how diverse sets of promoter recognition factors are utilized to match these core promoter elements. - PubMed
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. This work shows that the TAF3 subunit of TFIID can selectively bind to histone H3 trimethylated at lysine 4, and loss of this modification reduces TFIID binding to and transcription from a subset of promoters in vivo. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases