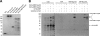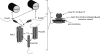Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance - PubMed (original) (raw)
Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance
Evgeny Onischenko et al. J Cell Biol. 2009.
Abstract
The nuclear pore complex (NPC) mediates all nucleocytoplasmic transport, yet its structure and biogenesis remain poorly understood. In this study, we have functionally characterized interaction partners of the yeast transmembrane nucleoporin Ndc1. Ndc1 forms a distinct complex with the transmembrane proteins Pom152 and Pom34 and two alternative complexes with the soluble nucleoporins Nup53 and Nup59, which in turn bind to Nup170 and Nup157. The transmembrane and soluble Ndc1-binding partners have redundant functions at the NPC, and disruption of both groups of interactions causes defects in Ndc1 targeting and in NPC structure accompanied by significant pore dilation. Using photoconvertible fluorescent protein fusions, we further show that the depletion of Pom34 in cells that lack NUP53 and NUP59 blocks new NPC assembly and leads to the reversible accumulation of newly made nucleoporins in cytoplasmic foci. Therefore, Ndc1 together with its interaction partners are collectively essential for the biosynthesis and structural integrity of yeast NPCs.
Figures
Figure 1.
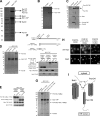
Identification of Ndc1-interacting proteins and characterization of the Ndc1 subcomplex. (A) Affinity purification from yeast expressing TAP-tagged Ndc1 (NDC1-TAP) or untagged controls. Gel areas were excised and subjected to LC-MS/MS. Lines mark borders of areas subjected to MS analysis. (B) Profile of an Ndc1 affinity purification performed in low ionic strength conditions. (C) Comparison of purifications performed from cells expressing TAP-tagged versions of Pom34 (POM34-TAP), Pom152 (POM152-TAP), or untagged controls. (D) Comparison of Ndc1-TAP affinity purifications from wild-type (wt), _pom34_Δ, _pom152_Δ, or untagged control cells. For A–D, purified proteins were eluted, separated by SDS-PAGE, and visualized by SYPRO-Ruby. (E) Western blot analysis of the levels of myc-tagged Pom152 or myc-tagged Pom34 in wild-type, _pom34_Δ, or _pom152_Δ cells expressing Ndc1-TAP. Ndc1-TAP and Xpo1/Crm1 levels are used as loading controls. (F) Western blot analysis of Ndc1 purifications from cells expressing Ndc1-TAP together with either 13×myc-tagged Pom152full-length, Pom152170–1337, or Pom1521–301. The scheme depicts the membrane topology of Pom152. Xpo1 was included as a control. sup, supernatant. (G) Comparison of Ndc1 purifications from cells expressing full-length Pom152-13×myc or its truncations. Proteins were visualized with SYPRO-Ruby. (H) Distribution of full-length Pom152 and its truncations tagged with 13×myc analyzed by immunofluorescence. Nuclei were stained with DAPI. (I) Model summarizing the interactions between Ndc1, Pom152, and Pom34. Solid arrows indicate direct protein interactions. The dashed arrow displays the functional requirement of Pom34 for the Ndc1 subcomplex integrity. Migration rate references for all gel image panels are shown in kilodaltons. Bar, 5 µm.
Figure 2.
The interactions of Ndc1 with Nup53 and Nup59. (A) Purified proteins separated by SDS-PAGE and stained with Coomassie brilliant blue. (B and C) SYPRO-Ruby–stained gels showing input (load) or eluates from S beads with immobilized Ndc1-TAP or S beads alone incubated with E. coli extract (bact. extract) or E. coli extract premixed with either GST-Nup53 or GST-Nup59. (D, top) Purified proteins stained with Coomassie brilliant blue. (bottom) SYPRO-Ruby–stained gel showing eluates from Ndc1-TAP beads after incubation with variants of Nup53 and Nup59. (E) Summary of interactions between truncations of Nup53 or Nup59 and Ndc1. (F) Binding of GST-Nup59 to Ndc1-TAP beads after preincubation with 10-fold excess of GST or 10-fold access of GST-Nup53. (G) Comparison of GST-Nup59 binding with Ndc1-TAP beads after preincubation with an excess of GST-Nup53 truncations or GST alone (left), or the reciprocal experiment with GST-Nup53 and an excess of GST-Nup59 truncations or GST alone (right). (H) Model summarizing the interactions between Ndc1 and Nup53 or Nup59. Solid arrows indicate direct protein interactions. Dashed arrow reflects the competition observed between Nup53 and Nup59 for Ndc1 binding. Migration rate references for all gel image panels are shown in kilodaltons.
Figure 3.
The interactions between Nup53, Nup59, Nup157, and Nup170. (A) Coomassie brilliant blue–stained gel showing samples of purified proteins. (B) SYPRO-Ruby–stained gel showing samples of the load (left) or eluates from TAP-Nup170, TAP-Nup157, or TAP beads after incubation with different mixtures of E. coli extract, 6×His-MBP-Nup53, 6×His-MBP-Nup59, or 6×His-MBP. Migration rate references for all gel image panels are shown in kilodaltons.
Figure 4.
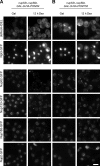
The role of Pom152, Pom34, Nup53, and Nup59 in targeting of Ndc1 to NE. (A) Localization of Ndc1 was compared in _nup53_Δ _pom34_Δ _pom152_Δ MET3-3×HA-NUP59 (i), _nup53_Δ MET3-3×HA-NUP59 (ii), _pom34_Δ _pom152_Δ GAL-3×HA-NUP59 (iii), and GAL-3×HA-NUP59 (iv) cells by fluorescence microscopy (via the yeast EGFP tag in i and ii) or by indirect immunofluorescence (via 13×myc tag in iii and iv) in permissive (galactose medium [gal] or medium lacking methionine [−met]) and nonpermissive conditions (dextrose-containing medium [dex] or medium containing 10× methionine [+10× met]). For immunofluorescence, cells were also stained with DAPI to label DNA. (B) Similar analysis of Ndc1–yeast EGFP localization in _nup53_Δ _nup59_Δ GAL-3×HA-POM34 (i), GAL-3×HA-POM34 (ii), _nup53_Δ _nup59_Δ GAL-3×HA-POM152 (iii), and GAL-3×HA-POM152 (iv). Bars, 5 µm.
Figure 5.
Roles of Nup53 and Nup59 in NPC organization and function. (A and B) _pom152_Δ _pom34_Δ GAL-3×HA-NUP59 (A) or _pom152_Δ _pom34_Δ _nup53_Δ MET3-3×HA-NUP59 (B) cells expressing GFP-tagged Nup82, Nup133, Nup188, or Nup2 or nuclear import reporters Npl3-GFP (B) or Npl3(S411A)-GFP (A) together with the ER reporter dsRed-HDEL were analyzed by fluorescence microscopy in both permissive and nonpermissive conditions. Gal, galactose medium; dex, dextrose-containing medium; −met, medium lacking methionine; +10× met, medium containing 10× methionine. Bar, 5 µm.
Figure 6.
Roles of Pom152 and Pom34 in NPC organization and function. (A and B) Fluorescence analysis as described in Fig. 5 in _nup53_Δ _nup59_Δ cells conditionally expressing Pom34 (A) or Pom152 (B). Gal, galactose medium; dex, dextrose-containing medium. Bar, 5 µm.
Figure 7.
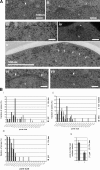
Transmission EM analysis of NPC morphology in conditional mutants affecting Ndc1-interacting nucleoporins. (A) NPCs (arrowheads) in _pom152_Δ _pom34_Δ GAL-3×HA-NUP59 cells in permissive conditions (i) did not display morphological abnormalities. (ii) After incubation in nonpermissive conditions, a large fraction of dilated pores often displaying fuzzy electron-dense material could be detected (asterisks). (iii) A similar phenotype was observed in _pom152_Δ _pom34_Δ _nup53_Δ MET3-3×HA-NUP59 cells in permissive conditions. After incubation in nonpermissive conditions, these cells displayed not only dilated pores (iv) but also large openings in NE (v, arrows). In _nup53_Δ _nup59_Δ GAL-3×HA-POM34 cells, no obvious NPC size abnormalities could be detected both in permissive (vi) and nonpermissive conditions (vii). n, nucleus. Bars: (i–iv, vi, and vii) 100 nm; (v) 500 nm. (B, i–iii) Comparison of pore size distributions in _pom152_Δ _pom34_Δ GAL-3×HA-NUP59 cells (i), _pom152_Δ _pom34_Δ _nup53_Δ MET3-3×HA-NUP59 cells (ii), and _nup53_Δ _nup59_Δ GAL-3×HA-POM34 cells (iii) incubated in permissive and nonpermissive conditions. (iv) Comparison of pore numbers per section in _nup53_Δ _nup59_Δ GAL-3×HA-POM34 in either permissive or nonpermissive conditions. Gal, galactose medium; dex, dextrose-containing medium; −met, medium lacking methionine; 10× met, medium containing 10× methionine. Error bars indicate SEM.
Figure 8.
Dendra assay to analyze distribution of nucleoporins in nup53Δ nup59Δ GAL-3×HA-POM34 cells. (A) Representation of the use of Nup-Dendra fusions to track the localization of newly synthesized and preexisting nucleoporins. Initially, Dendra displays green fluorescence but can be irreversibly photoconverted into a red fluorophore by a short pulse of UV light. After conversion, newly made nucleoporins are detected in the green channel, whereas red fluorescence can be used to localize Nups that were synthesized before the photoconversion. (B) Distribution of red and green Nup133-2×Dendra variants before (left), immediately after (middle), and 30 min after photoconversion (right). (C) Time course to visualize the Nup82-Dendra distribution in _nup53_Δ _nup59_Δ GAL-3×HA-POM34 cells after repression of POM34 expression without photoconversion. (D) Distribution of newly made and preexisting Nup82-Dendra in _nup53_Δ _nup59_Δ GAL-3×HA-POM34 cells. Cells were incubated for 4 h in nonpermissive conditions, photoconverted (left), and the same field of cells was reimaged after an additional 6-h incubation period (right). (E) Cells were incubated in dextrose medium (dex) for 9 h to induce Nup82 mislocalization, placed in galactose medium (gal), and photoconverted (left). After a 3-h incubation, the same cells were imaged again to detect the distribution of preexisting and newly synthesized Nup82-Dendra. Bars, 5 µm.
Figure 9.
Summary model of the Ndc1 interaction network. Solid arrows depict direct protein interactions. The dashed arrow displays the functional requirement of Pom34 for the Ndc1 subcomplex integrity.
Comment in
- Piecing together nuclear pore complex assembly during interphase.
Rexach M. Rexach M. J Cell Biol. 2009 May 4;185(3):377-9. doi: 10.1083/jcb.200904022. J Cell Biol. 2009. PMID: 19414605 Free PMC article.
Similar articles
- A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly.
Lau CK, Giddings TH Jr, Winey M. Lau CK, et al. Eukaryot Cell. 2004 Apr;3(2):447-58. doi: 10.1128/EC.3.2.447-458.2004. Eukaryot Cell. 2004. PMID: 15075274 Free PMC article. - The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function.
Madrid AS, Mancuso J, Cande WZ, Weis K. Madrid AS, et al. J Cell Biol. 2006 May 8;173(3):361-71. doi: 10.1083/jcb.200506199. J Cell Biol. 2006. PMID: 16682526 Free PMC article. - Distinct domains in Ndc1 mediate its interaction with the Nup84 complex and the nuclear membrane.
Amm I, Weberruss M, Hellwig A, Schwarz J, Tatarek-Nossol M, Lüchtenborg C, Kallas M, Brügger B, Hurt E, Antonin W. Amm I, et al. J Cell Biol. 2023 Jun 5;222(6):e202210059. doi: 10.1083/jcb.202210059. Epub 2023 May 8. J Cell Biol. 2023. PMID: 37154843 Free PMC article. - [Nuclear pores: from yeast to higher eukaryotes].
Doye V. Doye V. J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review.
Cited by
- 3D ultrastructure of the nuclear pore complex.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Curr Opin Cell Biol. 2012 Feb;24(1):86-91. doi: 10.1016/j.ceb.2011.12.011. Epub 2012 Jan 11. Curr Opin Cell Biol. 2012. PMID: 22244612 Free PMC article. Review. - Integrity and function of the Saccharomyces cerevisiae spindle pole body depends on connections between the membrane proteins Ndc1, Rtn1, and Yop1.
Casey AK, Dawson TR, Chen J, Friederichs JM, Jaspersen SL, Wente SR. Casey AK, et al. Genetics. 2012 Oct;192(2):441-55. doi: 10.1534/genetics.112.141465. Epub 2012 Jul 13. Genetics. 2012. PMID: 22798490 Free PMC article. - An amphipathic helix in Brl1 is required for nuclear pore complex biogenesis in S. cerevisiae.
Kralt A, Wojtynek M, Fischer JS, Agote-Aran A, Mancini R, Dultz E, Noor E, Uliana F, Tatarek-Nossol M, Antonin W, Onischenko E, Medalia O, Weis K. Kralt A, et al. Elife. 2022 Aug 24;11:e78385. doi: 10.7554/eLife.78385. Elife. 2022. PMID: 36000978 Free PMC article. - Identification of Saccharomyces cerevisiae spindle pole body remodeling factors.
Greenland KB, Ding H, Costanzo M, Boone C, Davis TN. Greenland KB, et al. PLoS One. 2010 Nov 12;5(11):e15426. doi: 10.1371/journal.pone.0015426. PLoS One. 2010. PMID: 21103054 Free PMC article. - Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes.
Hetzer MW, Wente SR. Hetzer MW, et al. Dev Cell. 2009 Nov;17(5):606-16. doi: 10.1016/j.devcel.2009.10.007. Dev Cell. 2009. PMID: 19922866 Free PMC article. Review.
References
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p.J. Cell Biol. 131:1133–1148 - PMC - PubMed
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007a. Determining the architectures of macromolecular assemblies.Nature. 450:683–694 - PubMed
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007b. The molecular architecture of the nuclear pore complex.Nature. 450:695–701 - PubMed
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I.W. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation.Mol. Cell. 17:83–92 - PubMed
- Antonin W., Ellenberg J., Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization.FEBS Lett. 582:2004–2016 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases