Single-molecule approaches to stochastic gene expression - PubMed (original) (raw)
Review
Single-molecule approaches to stochastic gene expression
Arjun Raj et al. Annu Rev Biophys. 2009.
Abstract
Both the transcription of mRNAs from genes and their subsequent translation into proteins are inherently stochastic biochemical events, and this randomness can lead to substantial cell-to-cell variability in mRNA and protein numbers in otherwise identical cells. Recently, a number of studies have greatly enhanced our understanding of stochastic processes in gene expression by utilizing new methods capable of counting individual mRNAs and proteins in cells. In this review, we examine the insights that these studies have yielded in the field of stochastic gene expression. In particular, we discuss how these studies have played in understanding the properties of bursts in gene expression. We also compare the array of different methods that have arisen for single mRNA and protein detection, highlighting their relative strengths and weaknesses. In conclusion, we point out further areas where single-molecule techniques applied to gene expression may lead to new discoveries.
Figures
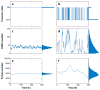
Figure 1
(a) Promoter dynamics for a gene that is always in the active state (i.e., nonbursting) versus (b) promoter dynamics for a gene that switches between active and inactive states (i.e., bursty dynamics). (c) mRNA dynamics for nonbursting and (d) bursting genes. In the nonbursting case, one obtains a Poisson distribution of mRNAs per cell across the population, as shown in the marginal histogram, whereas the distribution of mRNAs per cell in the bursting case is much wider than a Poisson distribution despite having the same mean. Protein dynamics for (e) nonbursting and (f) bursting genes, again with the same mean. Although the underlying gene expression dynamics are bursty, the relatively long half-life of the protein results in a wide but Gaussian-looking population distribution, pointing out the need for single-molecule mRNA-counting approaches when studying bursty gene expression. The marginal histograms on the right of the time courses show the distribution of the promoter states, mRNAs, and proteins across a population.
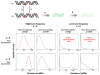
Figure 2
Distributions resulting from different values of the parameters in the gene activation/inactivation model of Peccoud & Ycart (28). The top row corresponds to the parameter γ being larger than the mRNA decay rate δ. The left side of the figure corresponds to high burst frequency compared with δ, whereas the right side corresponds to low burst frequency. The transcription rate γ was also altered as indicated. As mentioned in the text, the burst approximation is only valid when the burst frequency is low and the inactivation rate is faster than the mRNA decay rate. In particular, the bimodal expression pattern that appears with high μ and small λ, and γ cannot appear when one uses the burst approximation.
Similar articles
- Distribution of population-averaged observables in stochastic gene expression.
Bhattacharyya B, Kalay Z. Bhattacharyya B, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2014 Jan;89(1):012715. doi: 10.1103/PhysRevE.89.012715. Epub 2014 Jan 21. Phys Rev E Stat Nonlin Soft Matter Phys. 2014. PMID: 24580265 - Analytical distributions for detailed models of stochastic gene expression in eukaryotic cells.
Cao Z, Grima R. Cao Z, et al. Proc Natl Acad Sci U S A. 2020 Mar 3;117(9):4682-4692. doi: 10.1073/pnas.1910888117. Epub 2020 Feb 18. Proc Natl Acad Sci U S A. 2020. PMID: 32071224 Free PMC article. - A theoretical analysis of gene selection.
Mukherjee S, Roberts SJ. Mukherjee S, et al. Proc IEEE Comput Syst Bioinform Conf. 2004:131-41. Proc IEEE Comput Syst Bioinform Conf. 2004. PMID: 16448007 - Stochastic and delayed stochastic models of gene expression and regulation.
Ribeiro AS. Ribeiro AS. Math Biosci. 2010 Jan;223(1):1-11. doi: 10.1016/j.mbs.2009.10.007. Epub 2009 Oct 31. Math Biosci. 2010. PMID: 19883665 Review. - Microarray analysis of variation in individual aging C. elegans: approaches and challenges.
Golden TR, Hubbard A, Melov S. Golden TR, et al. Exp Gerontol. 2006 Oct;41(10):1040-5. doi: 10.1016/j.exger.2006.06.034. Epub 2006 Jul 28. Exp Gerontol. 2006. PMID: 16876364 Review.
Cited by
- Single-molecule RNA-FISH analysis reveals stochasticity in reactivation of latent HIV-1 regulated by Nuclear Orphan Receptors NR4A and cMYC.
LaPorte A, Pathak R, Eliscovich C, Martins L, Nell R, Spivak A, Suzuki M, Planelles V, Singer R, Kalpana G. LaPorte A, et al. Res Sq [Preprint]. 2024 Apr 19:rs.3.rs-4166090. doi: 10.21203/rs.3.rs-4166090/v1. Res Sq. 2024. PMID: 38699331 Free PMC article. Preprint. - miRNA circuit modules for precise, tunable control of gene expression.
Du R, Flynn MJ, Honsa M, Jungmann R, Elowitz MB. Du R, et al. bioRxiv [Preprint]. 2024 Mar 12:2024.03.12.583048. doi: 10.1101/2024.03.12.583048. bioRxiv. 2024. PMID: 38559239 Free PMC article. Preprint. - Intrinsically disordered proteins: Ensembles at the limits of Anfinsen's dogma.
Kulkarni P, Leite VBP, Roy S, Bhattacharyya S, Mohanty A, Achuthan S, Singh D, Appadurai R, Rangarajan G, Weninger K, Orban J, Srivastava A, Jolly MK, Onuchic JN, Uversky VN, Salgia R. Kulkarni P, et al. Biophys Rev (Melville). 2022 Mar 17;3(1):011306. doi: 10.1063/5.0080512. eCollection 2022 Mar. Biophys Rev (Melville). 2022. PMID: 38505224 Free PMC article. Review. - How do stochastic processes and genetic threshold effects explain incomplete penetrance and inform causal disease mechanisms?
Jenkins D. Jenkins D. Philos Trans R Soc Lond B Biol Sci. 2024 Apr 22;379(1900):20230045. doi: 10.1098/rstb.2023.0045. Epub 2024 Mar 4. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38432317 Free PMC article. Review. - On the ever-growing functional versatility of the CRISPR-Cas13 system.
Montagud-Martínez R, Márquez-Costa R, Heras-Hernández M, Dolcemascolo R, Rodrigo G. Montagud-Martínez R, et al. Microb Biotechnol. 2024 Feb;17(2):e14418. doi: 10.1111/1751-7915.14418. Microb Biotechnol. 2024. PMID: 38381083 Free PMC article. Review.
References
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–32. - PubMed
- Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–75. - PubMed
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. - PubMed
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–78. - PubMed
Publication types
MeSH terms
Grants and funding
- R01-GM077183/GM/NIGMS NIH HHS/United States
- R01 GM077183-03/GM/NIGMS NIH HHS/United States
- R01-GM068957/GM/NIGMS NIH HHS/United States
- R01 GM068957-07/GM/NIGMS NIH HHS/United States
- R01 GM077183/GM/NIGMS NIH HHS/United States
- R01 GM068957/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources