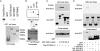SUMO interaction motifs in Sizn1 are required for promyelocytic leukemia protein nuclear body localization and for transcriptional activation - PubMed (original) (raw)
SUMO interaction motifs in Sizn1 are required for promyelocytic leukemia protein nuclear body localization and for transcriptional activation
Ginam Cho et al. J Biol Chem. 2009.
Abstract
Mutations in Sizn1 (Zcchc12), a novel transcriptional co-activator in the BMP signaling pathway, are associated with X-linked mental retardation. Previously, we demonstrated that Sizn1 positively modulates the BMP signal by interacting with Smad family members and cAMP-responsive element-binding protein-binding protein. To further define the molecular basis of Sizn1 function, we have explored its subcellular localization and generated various deletion mutants to carry out domain analyses. Here, we report that Sizn1 localizes to promyelocytic leukemia protein nuclear bodies (PML-NBs). Sizn1 deletion mutants that disrupt the MA homologous domain or the middle region fail to target to the PML-NB. We show that two SUMO interaction motifs (SIMs) in Sizn1 can bind to SUMO and govern SUMO conjugation to Sizn1 in the absence of the consensus motif for SUMO attachment. Interestingly, the SIM mutant Sizn1 localizes to nuclear bodies, but not to PML-NBs. Thus, SIMs mediate the localization of Sizn1 to PML-NB. Interestingly, mutations in SIM sequences and deletion of the MA homologous domain also affected the transcriptional co-activation function of a Sizn1. Taken together, our data indicate that the SIMs in Sizn1 are required for its PML-NB localization and for the full transcriptional co-activation function in BMP signaling.
Figures
FIGURE 1.
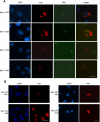
Sizn1 is a nuclear protein localized in PML bodies. A, the GFP-Sizn1 fusion protein is localized in a speckled pattern within the nucleus of HEK293T cells. B, endogenous Sizn1, detected by immunofluorescence using an anti-Sizn1 antibody, is expressed in a nuclear speckle pattern in the septal nucleus of the P3 neonatal brain. (Lower left box is a high power image corresponding to the smaller box in each image.) C, Sizn1 co-localizes with nuclear PML bodies in C2C12 cells that were transfected with pMIWIII/Myc-Sizn1. Scale bars indicate 10 μm.
FIGURE 2.
Schematic representation of Sizn1 functional domains and deletion mutants used in this study. Sizn1 has an MA homologous domain at the N terminus, two SIMs, and a putative NLS. The Myc tag is illustrated with a purple box, and the GFP tag is illustrated with a green box. N, nucleus; C, cytoplasm. Boldface and italic type on SIM represent hydrophobic and positively charged amino acid residues, respectively. Arrows indicate mutated amino acids in SIM2 and SIM3 (Val, Ile to Ala, Ala). The putative NLS region (200–250) identified in this study is not depicted in the diagram. The constructs used for in vitro translation did not contain the Myc tag. WT, wild type.
FIGURE 3.
Subcellular localization of Sizn1 deletion mutants tagged with Myc. C2C12 cells were transfected with the indicated deletion mutants and immunostained with anti-PML and anti-Myc. A, C-terminal deletion mutants. Only Myc-1–345, Sizn1 mutants containing the NLS. B, _N_-terminal deletion mutants. Only the Myc-32–402 mutant showed proper PML-NB localization, whereas the others showed diffused nuclear distribution. In all figures, 4′,6-diamidino-2-phenylindole (DAPI) is used to stain the nucleus. Scale bars indicate 10 μm.
FIGURE 4.
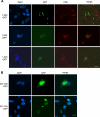
Subcellular localization of GFP-tagged Sizn1 deletion mutants. Immunostaining of GFP-tagged mutants in transfected C2C12 cells using anti-PML. A, C-terminal deletion mutants. GFP fluorescence merged with PML antibody staining shows that only the 1–345-GFP is correctly targeted to the PML-NB. B, deletion mutants containing an NLS (predicted and unpredicted). 321–345 (predicted NLS) can target GFP to the nucleus but not to the PML-NB. 200–320 (without predicted NLS) can target GFP to the nucleus with weak cytoplasmic distribution. Scale bar indicates 10 μm. DAPI, 4′,6-diamidino-2-phenylindole.
FIGURE 5.
Sizn1 is a SUMOylated protein, and its SUMOylation is dependent on SIMs. A, GST/SUMO pulldown assay with HEK293T cells transfected with GST-SUMO and Myc-Sizn1 plasmids (full-length and deletion mutants). Both SUMOylated (covalent conjugation, white arrow) and nonSUMOylated forms of Sizn1 (noncovalent interaction with SUMO, black arrow) are detected when the wild type (WT) and Sizn1 mutants were co-transfected with the exception of the Myc-1–250 mutant. Note that the C-terminal deletion mutants migrate slowly in SDS-PAGE for their size. B, autoradiography of the GST/SUMO pulldown assay with _in vitro_-translated and 35S-labeled Sizn1. The mSIM1, mSIM2, mSIM3, and mSIM4 contain two amino acid mutations in a single SIM sequence, whereas mSIM2/3 harbors mutations in both SIM2 and -3. C, GST pulldown assay with HEK293T cells transfected with GST-SUMO and Sizn1 (the wild type and mutants contain modified SIMs). The level of both SUMOylated and nonSUMOylated forms of Sizn1 are pulled down, but reduced levels are found for mSIM2; none are detected with the mSIM3 and mSIM2/3 mutants.
FIGURE 6.
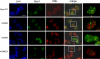
Mutations in SIM sequences cause Sizn1 to localize in distinct nuclear puncta that do not overlap with PML-NB. C2C12 cells were transfected with the Sizn1 wild type (WT) and mutant forms followed by immunostaining with anti-PML and anti-Sizn1 antibodies. In mutants, speckled staining of the Sizn1 does not overlap with PML-NB, unlike in the wild type. The right column identifies the enlarged images in the merged image. Scale bar indicates 10 μm. DAPI, 4′,6-diamidino-2-phenylindole.
FIGURE 7.
Sizn1 maintains punctate localization in the nucleus in the absence of PML protein. C2C12 cells were transfected with PML-shRNA (GFP) and Sizn1 expression constructs. The cells were immuno-labeled with anti-PML (blue color) and anti-Sizn1(red color) antibodies. Cells transfected with the PML-shRNA expression construct show GFP (green, expressed from vector) but no PML staining. However, punctate labeling for Sizn1 is still present. Arrows indicate the PML-shRNA expressing cells. The solid and dashed circles indicate nontransfected and transfected nuclei, respectively. Scale bar indicates 10 μm.
FIGURE 8.
The C terminus of Sizn1 interacts with the MH1 domain of Smad1 and the N terminus of CBP. A and B, autoradiography of the in vitro GST pulldown assay. A, GST-Smad1(full-length), GST-MH1 (only MH1 domain of the Smad1), and GST-MH2 (MH2 domain of the Smad1) were used to pull down _in vitro_-translated 35S-labeled Sizn1 (full-length). B, GST-Smad1 was used to pull down _in vitro_-translated 35S-labeled Sizn1 (full-length, 1–200, and 201–402, respectively). Full-length Sizn1 and the 201–402 mutant protein interact with Smad1. The 1–200 mutant protein showed minimal signals very similar to the negative control. C and D, GST pulldown assay with HEK293T cells. C, full-length Sizn1 and various GST-CBP deletion constructs were co-transfected. Only CBP(N) and 1–770 are able to pull down Sizn1. N, 770, KIX, and C indicate 1–450, 1–770, the KIX domain, and 1891–2441 fused with GST, respectively. D, GST-CBP(N) and various Sizn1 deletion mutants were co-transfected. Only 1–250-GFP (Sizn1) is pulled down with GST-CBP(N).
FIGURE 9.
PML-NB localization of Sizn1 is important for its functional co-activator of the BMP signaling pathway. _SBEx4_-luciferase and caBMPR1a were co-transfected with Myc-tagged Sizn1 deletion mutants (A) or SIM point mutants (B) into C2C12 cells, and luciferase activities were measured (n = 4 and n = 2, respectively). Activity is diminished in the presence of Sizn1 mutant protein (p < 0.05 for 1–200, 1–250, 201–402, 251–402, and 321–402-GFP compared with control (WT) (Student's t test)). No statistical difference was found for the 1–345 and 1–380 constructs (p < 0.05 for mSIM1, mSIM2, mSIM3, and mSIM2/3; compare with control (WT)). Error bars indicate the S.D.
Similar articles
- Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3.
Fu C, Ahmed K, Ding H, Ding X, Lan J, Yang Z, Miao Y, Zhu Y, Shi Y, Zhu J, Huang H, Yao X. Fu C, et al. Oncogene. 2005 Aug 18;24(35):5401-13. doi: 10.1038/sj.onc.1208714. Oncogene. 2005. PMID: 15940266 - Disruption of PML nuclear bodies is mediated by ORF61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin.
Wang L, Oliver SL, Sommer M, Rajamani J, Reichelt M, Arvin AM. Wang L, et al. PLoS Pathog. 2011 Aug;7(8):e1002157. doi: 10.1371/journal.ppat.1002157. Epub 2011 Aug 25. PLoS Pathog. 2011. PMID: 21901090 Free PMC article. - Role of SUMO in RNF4-mediated promyelocytic leukemia protein (PML) degradation: sumoylation of PML and phospho-switch control of its SUMO binding domain dissected in living cells.
Percherancier Y, Germain-Desprez D, Galisson F, Mascle XH, Dianoux L, Estephan P, Chelbi-Alix MK, Aubry M. Percherancier Y, et al. J Biol Chem. 2009 Jun 12;284(24):16595-16608. doi: 10.1074/jbc.M109.006387. Epub 2009 Apr 20. J Biol Chem. 2009. PMID: 19380586 Free PMC article. - Role of nuclear bodies in apoptosis signalling.
Krieghoff-Henning E, Hofmann TG. Krieghoff-Henning E, et al. Biochim Biophys Acta. 2008 Nov;1783(11):2185-94. doi: 10.1016/j.bbamcr.2008.07.002. Epub 2008 Jul 16. Biochim Biophys Acta. 2008. PMID: 18680765 Review. - A Tale of Usurpation and Subversion: SUMO-Dependent Integrity of Promyelocytic Leukemia Nuclear Bodies at the Crossroad of Infection and Immunity.
Patra U, Müller S. Patra U, et al. Front Cell Dev Biol. 2021 Aug 27;9:696234. doi: 10.3389/fcell.2021.696234. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513832 Free PMC article. Review.
Cited by
- A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Krüppel-like factor 4.
Du JX, McConnell BB, Yang VW. Du JX, et al. J Biol Chem. 2010 Sep 3;285(36):28298-308. doi: 10.1074/jbc.M110.101717. Epub 2010 Jun 28. J Biol Chem. 2010. PMID: 20584900 Free PMC article. - The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism.
Wang Y, Yu Y, Pang Y, Yu H, Zhang W, Zhao X, Yu J. Wang Y, et al. RNA Biol. 2021 Dec;18(12):2107-2126. doi: 10.1080/15476286.2021.1909320. Epub 2021 May 4. RNA Biol. 2021. PMID: 33787465 Free PMC article. Review. - PML, SUMOylation, and Senescence.
Ivanschitz L, De Thé H, Le Bras M. Ivanschitz L, et al. Front Oncol. 2013 Jul 4;3:171. doi: 10.3389/fonc.2013.00171. eCollection 2013. Front Oncol. 2013. PMID: 23847762 Free PMC article. - Regulation of Cellular Ribonucleoprotein Granules: From Assembly to Degradation via Post-translational Modification.
Jeon P, Ham HJ, Park S, Lee JA. Jeon P, et al. Cells. 2022 Jun 29;11(13):2063. doi: 10.3390/cells11132063. Cells. 2022. PMID: 35805146 Free PMC article. Review. - ZCCHC12, a potential molecular marker of papillary thyroid carcinoma: a preliminary study.
Li QL, Chen FJ, Lai R, Guo ZM, Luo R, Yang AK. Li QL, et al. Med Oncol. 2012 Sep;29(3):1409-17. doi: 10.1007/s12032-011-0018-6. Epub 2011 Jul 8. Med Oncol. 2012. PMID: 21739307
References
- Caspary T., Anderson K. V. (2003) Nat. Rev. Neurosci. 4, 289–297 - PubMed
- Hsieh J., Gage F. H. (2005) Curr. Opin. Cell Biol. 17, 664–671 - PubMed
- López-Coviella I., Berse B., Krauss R., Thies R. S., Blusztajn J. K. (2000) Science 289, 313–316 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous