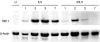Sleeping beauty transposition from nonintegrating lentivirus - PubMed (original) (raw)
Sleeping beauty transposition from nonintegrating lentivirus
Conrad A Vink et al. Mol Ther. 2009 Jul.
Abstract
Lentiviral vectors enter cells with high efficiency and deliver stable transduction through integration into host chromosomes, but their preference for integration within actively transcribing genes means that insertional mutagenesis following disruption of host proto-oncogenes is a recognized concern. We have addressed this problem by combining the efficient cell and nuclear entry properties of HIV-1-derived lentiviral vectors with the integration profile benefits of Sleeping Beauty (SB) transposase. Importantly, this integration enzyme does not exhibit a preference for integration within active genes. We generated integrase-deficient lentiviral vectors (IDLVs) to carry SB transposon and transposase expression cassettes. IDLVs were able to deliver transient transposase expression to target cells, and episomal lentiviral DNA was found to be a suitable substrate for integration via the SB pathway. The hybrid vector system allows genomic integration of a minimal promoter-transgene cassette flanked by short SB inverted repeats (IRs) but devoid of HIV-1 long terminal repeats (LTRs) or other virus-derived sequences. Importantly, integration site analysis revealed redirection toward a profile mimicking SB-plasmid integration and away from integration within transcriptionally active genes favored by integrase-proficient lentiviral vectors (ILVs).
Figures
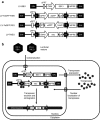
**Figure 1
Constructs and experimental strategy. (a) Lentiviral vector constructs. LV-SB11 carries a Sleeping Beauty transposase expression cassette. LV-TeGFP FWD and LV-TeGFP REV carry a Sleeping Beauty transposon containing an eGFP expression cassette that is in the sense and antisense orientations respectively. LV-TNEO carries a Sleeping Beauty transposon containing a neomycin phosphotransferase expression cassette in the sense orientation. CMV, immediate early promoter of human cytomegalovirus; cPPT, central polypurine tract; eGFP, enhanced green fluorescent protein; FWD, forward; IL, Sleeping Beauty transposon left inverted repeat; IR, Sleeping Beauty transposon right inverted repeat; LV, lentiviral vector; neo, neomycin-phosphotransferase gene; pA, SV40 polyadenylation signal; REV, reverse; SB11, hyperactive Sleeping Beauty transposase; SFFV; spleen focus forming virus LTR promoter; SIN, self-inactivating (U3-deleted) HIV-1 long terminal repeat; SV40, simian virus 40 promoter; TeGFP, transposon with enhanced green fluorescent protein; TNEO, transposon with neomycin resistance; WPRE, woodchuck posttranscriptional regulatory element. (b) Experimental strategy used in this study. Cells are cotransduced with integrase-deficient lentiviral vectors carrying the Sleeping Beauty transposase and transposon. Transposase protein is expressed and localized to the nucleus where it binds to the transposon inverted repeats and catalyzes excision of the transposon from episomal lentiviral DNA. The excised transposon is mobile and able to subsequently reintegrate elsewhere, for example, into a host cell chromosome.
**Figure 2
Western blot for expression of transposase from lentiviral vectors. Integrase-proficient ILV-SB11 and integrase-deficient IDLV-SB11 transposase expression vectors were prepared in parallel and concentrated by ultracentrifugation. 106 HeLa cells were transduced with 0.5 µg p24 of vector per well. At 1, 2, 3, and 7 days post-transduction, cells were trypsinized and pellets of equal cell number were frozen for subsequent determination of protein expression by western blot. IDLV, integrase-deficient lentiviral vector; ILV, integrase-proficient lentiviral vector; U, untransduced.
**Figure 3
Efficiency of chromosomal integration. 105 HeLa cells were transduced or transfected with Sleeping Beauty components in a double titration integration assay. The transposon amounts tested are given by the y-axis labels in a, c, and e, whereas the transposase amounts are given by the x-axis labels. All transposon-transposase combinations were tested, and each combination was tested in triplicate. (a) Plasmid transposon, plasmid transposase; (b) cross-section at transposon plasmid mass was 4 µg; (c) plasmid transposon, IDLV transposase; (d) cross-section at transposon plasmid mass was 2 µg; (e) IDLV transposon, IDLV transposase; (f) cross-section where the SB-IDLV vector dose was 1.2 µg p24. The rate of integration was assessed by the number of G418-resistant colonies formed and is expressed as a percentage of the transduced or transfected cell number assuming 100% plating efficiency. IDLV, integrase-deficient lentiviral vector; SB11, hyperactive Sleeping Beauty transposase.
**Figure 4
Primary sequence at integration sites. (a) Sample junction sequences of integration sites recovered by ligation-mediated PCR. The beginning of the flanking chromosomal sequence is shown for illustration. Virus LTR or transposon IR sequence is shown in bold, and flanking chromosomal DNA is shown in normal type. (b) A sequence logo was generated using the WebLogo tool to identify preferred base pair usage at the three integration site types. Position 0 denotes the first base of the flanking chromosomal sequence 3′ of the integration site. ILV, integrating lentiviral vector; SB-IDLV, hybrid Sleeping Beauty–integrase-deficient lentiviral vector; SB-plasmid, Sleeping Beauty plasmid vector.
**Figure 5
Integration profiles of vector types. 105 HeLa cells per well were transduced with ILV-TNEO at multiplicity of infection 0.01 (ILV sites, top line), transduced with 1.2 µg p24 IDLV-TNEO and 0.13 µg IDLV-SB11 (SB-IDLV sites, middle line), or transfected with 4 µg pLV-TNEO and 1 µg pLV-SB11 (SB-plasmid sites, bottom line). Cells were incubated in nonselective medium for 3 days followed by a two-week incubation in medium containing 1 mg/ml G418. Genomic DNA was extracted and transposon-chromosome or lentivirus-chromosome junctions were recovered by ligation-mediated PCR and sequenced. (a) Sequences were mapped to the University of California at Santa Cruz (UCSC) human genome by BLAT search and integration sites were depicted relative to chromosomes using the UCSC Genome Graphs tool. Note that HeLa cells are karyotypically abnormal. (b) Intragenic position of integration sites within genes. RefSeq genes containing integration sites were divided by length into 10 equally sized regions and a 5 kb upstream region, and the proportion of integration sites within each region was counted. To allow statistical comparison of integration preferences with average genomic content, 1,000 random chromosomal sites were generated by multiplying the total length of the genome by a random number between 0 and 1 and converting this value to a chromosomal coordinate. Vector integration frequencies are expressed relative to the proportion of random sites within each region. (c) Transcriptional activity of genes containing integration sites. All RefSeq genes were scored for transcription in HeLa cells using a published microarray dataset. All genes were then assigned to one of three transcription levels (containing equal numbers of genes) to give low, medium, and highly transcribed genes. Integration sites within genes were then scored according to whether the hit gene was transcribed at a low, medium, or high level. For each vector type, the number of intragenic sites per transcription level is expressed as a percentage of the total number of intragenic sites. A dashed line at 33.3% of sites is included to show theoretically equal distribution of sites between the transcription levels. ILV, integrating lentiviral vector; SB-IDLV, hybrid Sleeping Beauty–integrase-deficient lentiviral vector; TNEO, transposon with neomycin resistance.
Similar articles
- Hybrid lentivirus-transposon vectors with a random integration profile in human cells.
Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z, Izsvák Z, Mikkelsen JG. Staunstrup NH, et al. Mol Ther. 2009 Jul;17(7):1205-14. doi: 10.1038/mt.2009.10. Epub 2009 Feb 24. Mol Ther. 2009. PMID: 19240688 Free PMC article. - Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells.
Moldt B, Miskey C, Staunstrup NH, Gogol-Döring A, Bak RO, Sharma N, Mátés L, Izsvák Z, Chen W, Ivics Z, Mikkelsen JG. Moldt B, et al. Mol Ther. 2011 Aug;19(8):1499-510. doi: 10.1038/mt.2011.47. Epub 2011 Apr 5. Mol Ther. 2011. PMID: 21468003 Free PMC article. - Hybrid adeno-associated viral vectors utilizing transposase-mediated somatic integration for stable transgene expression in human cells.
Zhang W, Solanki M, Müther N, Ebel M, Wang J, Sun C, Izsvak Z, Ehrhardt A. Zhang W, et al. PLoS One. 2013 Oct 8;8(10):e76771. doi: 10.1371/journal.pone.0076771. eCollection 2013. PLoS One. 2013. PMID: 24116154 Free PMC article. - Integrase-defective lentiviral vectors--a stage for nonviral integration machineries.
Staunstrup NH, Mikkelsen JG. Staunstrup NH, et al. Curr Gene Ther. 2011 Oct;11(5):350-62. doi: 10.2174/156652311797415881. Curr Gene Ther. 2011. PMID: 21745178 Review. - Sleeping Beauty Transposition.
Ivics Z, Izsvák Z. Ivics Z, et al. Microbiol Spectr. 2015 Apr;3(2):MDNA3-0042-2014. doi: 10.1128/microbiolspec.MDNA3-0042-2014. Microbiol Spectr. 2015. PMID: 26104705 Review.
Cited by
- Episomes and Transposases-Utilities to Maintain Transgene Expression from Nonviral Vectors.
Kreppel F, Hagedorn C. Kreppel F, et al. Genes (Basel). 2022 Oct 16;13(10):1872. doi: 10.3390/genes13101872. Genes (Basel). 2022. PMID: 36292757 Free PMC article. Review. - Delivering genes with human immunodeficiency virus-derived vehicles: still state-of-the-art after 25 years.
Wolff JH, Mikkelsen JG. Wolff JH, et al. J Biomed Sci. 2022 Oct 9;29(1):79. doi: 10.1186/s12929-022-00865-4. J Biomed Sci. 2022. PMID: 36209077 Free PMC article. Review. - Integrase deficient lentiviral vector: prospects for safe clinical applications.
Yew CT, Gurumoorthy N, Nordin F, Tye GJ, Wan Kamarul Zaman WS, Tan JJ, Ng MH. Yew CT, et al. PeerJ. 2022 Aug 12;10:e13704. doi: 10.7717/peerj.13704. eCollection 2022. PeerJ. 2022. PMID: 35979475 Free PMC article. - The Old and the New: Prospects for Non-Integrating Lentiviral Vector Technology.
Luis A. Luis A. Viruses. 2020 Sep 29;12(10):1103. doi: 10.3390/v12101103. Viruses. 2020. PMID: 33003492 Free PMC article. Review. - Preclinical and clinical advances in transposon-based gene therapy.
Tipanee J, Chai YC, VandenDriessche T, Chuah MK. Tipanee J, et al. Biosci Rep. 2017 Dec 5;37(6):BSR20160614. doi: 10.1042/BSR20160614. Print 2017 Dec 22. Biosci Rep. 2017. PMID: 29089466 Free PMC article. Review.
References
- Ivics Z, Hackett PB, Plasterk RH., and , Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. - PubMed
- Izsvák Z, Khare D, Behlke J, Heinemann U, Plasterk RH., and , Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–34588. - PubMed
- Cui Z, Geurts AM, Liu G, Kaufman CD., and , Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–1235. - PubMed
- Izsvák Z, Stüwe EE, Fiedler D, Katzer A, Jeggo PA., and , Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell. 2004;13:279–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources