High-throughput genotyping by whole-genome resequencing - PubMed (original) (raw)
. 2009 Jun;19(6):1068-76.
doi: 10.1101/gr.089516.108. Epub 2009 May 6.
Qi Feng, Qian Qian, Qiang Zhao, Lu Wang, Ahong Wang, Jianping Guan, Danlin Fan, Qijun Weng, Tao Huang, Guojun Dong, Tao Sang, Bin Han
Affiliations
- PMID: 19420380
- PMCID: PMC2694477
- DOI: 10.1101/gr.089516.108
High-throughput genotyping by whole-genome resequencing
Xuehui Huang et al. Genome Res. 2009 Jun.
Abstract
The next-generation sequencing technology coupled with the growing number of genome sequences opens the opportunity to redesign genotyping strategies for more effective genetic mapping and genome analysis. We have developed a high-throughput method for genotyping recombinant populations utilizing whole-genome resequencing data generated by the Illumina Genome Analyzer. A sliding window approach is designed to collectively examine genome-wide single nucleotide polymorphisms for genotype calling and recombination breakpoint determination. Using this method, we constructed a genetic map for 150 rice recombinant inbred lines with an expected genotype calling accuracy of 99.94% and a resolution of recombination breakpoints within an average of 40 kb. In comparison to the genetic map constructed with 287 PCR-based markers for the rice population, the sequencing-based method was approximately 20x faster in data collection and 35x more precise in recombination breakpoint determination. Using the sequencing-based genetic map, we located a quantitative trait locus of large effect on plant height in a 100-kb region containing the rice "green revolution" gene. Through computer simulation, we demonstrate that the method is robust for different types of mapping populations derived from organisms with variable quality of genome sequences and is feasible for organisms with large genome sizes and low polymorphisms. With continuous advances in sequencing technologies, this genome-based method may replace the conventional marker-based genotyping approach to provide a powerful tool for large-scale gene discovery and for addressing a wide range of biological questions.
Figures
Figure 1.
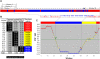
Sequence-based high-throughput genotyping. Rice RILs were developed from a cross between indica and japonica cultivars. Genome sequences of the parents were aligned and SNPs were identified. Genomes of the RILs were resequenced on the Illumina Genome Analyzer using the multiplexed sequencing strategy. Three-base indexed DNAs of 16 RILs were combined and sequenced in one lane. Sequences were sorted and aligned with the pseudomolecules of parental genome sequences for SNP detection. Detected SNPs were arranged along chromosomes according to their physical locations with genotypes indicated. A sliding window approach was used for genotype calling, recombination breakpoint determination, and map construction.
Figure 2.
Sliding window approach for genotype calling and recombination breakpoint determination. (A) The top stripe of blocks represents SNPs along the hypothetical chromosomal region. This was redrawn from the two stripes of short vertical lines below illustrating SNPs detected by aligning 33-mers with the parental genome sequences. (Red) Indica genotype; (blue) japonica genotype. A sliding window covering 15 SNPs moves from left to right one base at a time. For each window, the ratio of the number of indica to japonica SNPs (ind:jap) is calculated. (B) Genotype calling based on the highest expected probabilities: Call homozygous indica genotype (ind/ind) when ind:jap ≥ 11:4; call heterozygous genotype (ind/jap) when 10:5 ≥ ind:jap ≥ 3:12; call homozygous japonica genotype (jap/jap) when ind:jap ≤ 2:13. Adding together the probabilities of these callings (shaded in black) gives the calling accuracy of 99.94%. (C) As the window slides, genotypes are called and recombination breakpoints are determined. Green and brown arrows point to breakpoints between two homozygous genotypes and between the heterozygous and homozygous genotypes, respectively. The resulting recombination map for this chromosomal region is illustrated in a solid bar, in which red, blue, and yellow represent genotypes ind/ind, jap/jap, and ind/jap, respectively. Identified breakpoints are indicated between SNPs.
Figure 3.
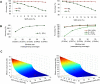
Simulation of genotype calling accuracy. (A) Effect of parental genome sequence quality on calling accuracy. (Left) One parent has high-quality genome sequences that give an SNP error rate of 1%, while the genome sequence quality of the other parent is allowed to vary and gives SNP error rates from 2% to 20%. (Right) Genome sequence qualities of both parents are allowed to vary and give the same SNP error rates from 2% to 20%. Two types of populations, RIL and F2, are considered, with ratios of three genotypes set at 49.5:1:49.5 and 1:2:1, respectively. Window size is set at 15. Genotype calling accuracy is calculated according to Equation 8 in Methods. (B) The effect of window size on calling accuracy. (Left) The critical error rate of 6% that drops the calling accuracy of F2 below 99% in the above figure is used. (Right) Three critical error rates are used, including 16% for both parents that drops the calling accuracy of RIL below 99%, 4% for both parents that drops the accuracy of F2 below 99%, and 12% for both parents that drops the accuracy of F2 below 95%, in the above figure. When window sizes are measured by the number of SNPs covering the same physical distance, increase in window sizes is equivalent to the increase in resequencing coverage. Rice is taken as an example to show resequencing coverage for the corresponding window size. (C) The amount of effective sequences (Se) required for a RIL to reach a range of mapping resolutions (R) as SNP densities (D) vary. (Left) Simulation for the rice genome size, 389 Mb. Red dot indicates the location of the rice RIL of this study (D = 3.2 SNPs/kb, R = 25 SNPs/Mb). (Right) Simulation for the mouse genome size, 2500 Mb. Red dot indicates Se required for a mouse RIL with D = 1.3 and R = 25.
Figure 4.
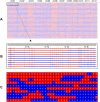
Recombination and bin maps. (A) Aligned recombination maps of 150 rice RILs. Red, ind/ind; blue, jap/jap; yellow, ind/jap. (B) Aligned chromosome 1 of the first ten RILs. Scale indicates physical distance. A vertical line labels a recombination breakpoint. A region between two vertical lines across all RILs is recognized as a recombination bin. (C) Bin map of the 10 RILs.
Similar articles
- Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.).
Xu J, Zhao Q, Du P, Xu C, Wang B, Feng Q, Liu Q, Tang S, Gu M, Han B, Liang G. Xu J, et al. BMC Genomics. 2010 Nov 24;11:656. doi: 10.1186/1471-2164-11-656. BMC Genomics. 2010. PMID: 21106060 Free PMC article. - Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum.).
Zhang Z, Shang H, Shi Y, Huang L, Li J, Ge Q, Gong J, Liu A, Chen T, Wang D, Wang Y, Palanga KK, Muhammad J, Li W, Lu Q, Deng X, Tan Y, Song W, Cai J, Li P, Rashid Ho, Gong W, Yuan Y. Zhang Z, et al. BMC Plant Biol. 2016 Apr 11;16:79. doi: 10.1186/s12870-016-0741-4. BMC Plant Biol. 2016. PMID: 27067834 Free PMC article. - Genetic Dissection of Germinability under Low Temperature by Building a Resequencing Linkage Map in japonica Rice.
Jiang S, Yang C, Xu Q, Wang L, Yang X, Song X, Wang J, Zhang X, Li B, Li H, Li Z, Li W. Jiang S, et al. Int J Mol Sci. 2020 Feb 14;21(4):1284. doi: 10.3390/ijms21041284. Int J Mol Sci. 2020. PMID: 32074988 Free PMC article. - Resequencing rice genomes: an emerging new era of rice genomics.
Huang X, Lu T, Han B. Huang X, et al. Trends Genet. 2013 Apr;29(4):225-32. doi: 10.1016/j.tig.2012.12.001. Epub 2013 Jan 4. Trends Genet. 2013. PMID: 23295340 Review. - Skim sequencing: an advanced NGS technology for crop improvement.
Kumar P, Choudhary M, Jat BS, Kumar B, Singh V, Kumar V, Singla D, Rakshit S. Kumar P, et al. J Genet. 2021;100:38. J Genet. 2021. PMID: 34238778 Review.
Cited by
- Genetic dissection of Meloidogyne incognita resistance genes based on VIGS functional analysis in Cucumis metuliferus.
Xie X, Ling J, Lu J, Mao Z, Zhao J, Zheng S, Yang Q, Li Y, Visser RGF, Bai Y, Xie B. Xie X, et al. BMC Plant Biol. 2024 Oct 15;24(1):964. doi: 10.1186/s12870-024-05681-6. BMC Plant Biol. 2024. PMID: 39402446 Free PMC article. - High-density genetic map construction and QTL mapping to identify genes for blight defense- and yield-related traits in sesame (Sesamum indicum L.).
Xu G, Cui Y, Li S, Guan Z, Miao H, Guo Y. Xu G, et al. Front Plant Sci. 2024 Sep 26;15:1446062. doi: 10.3389/fpls.2024.1446062. eCollection 2024. Front Plant Sci. 2024. PMID: 39391773 Free PMC article. - Integrative multi-omics analysis reveals genetic and heterotic contributions to male fertility and yield in potato.
Li D, Geng Z, Xia S, Feng H, Jiang X, Du H, Wang P, Lian Q, Zhu Y, Jia Y, Zhou Y, Wu Y, Huang C, Zhu G, Shang Y, Li H, Städler T, Yang W, Huang S, Zhang C. Li D, et al. Nat Commun. 2024 Oct 5;15(1):8652. doi: 10.1038/s41467-024-53044-4. Nat Commun. 2024. PMID: 39368981 Free PMC article. - Non-additive expression genes play a critical role in leaf vein ratio heterosis in Nicotiana tabacum L.
Duan L, Mo Z, Li K, Pi K, Luo J, Que Y, Zhang Q, Zhang J, Wu G, Liu R. Duan L, et al. BMC Genomics. 2024 Oct 3;25(1):924. doi: 10.1186/s12864-024-10821-1. BMC Genomics. 2024. PMID: 39363277 Free PMC article. - An ultra-dense linkage map identified quantitative trait loci corresponding to fruit quality- and size-related traits in red goji berry.
Rehman F, Gong H, Ma Y, Zeng S, Ke D, Yang C, Zhao Y, Wang Y. Rehman F, et al. Front Plant Sci. 2024 Sep 4;15:1390936. doi: 10.3389/fpls.2024.1390936. eCollection 2024. Front Plant Sci. 2024. PMID: 39297015 Free PMC article.
References
- Frazer K.A., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. - PubMed
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources