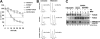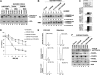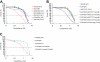Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M - PubMed (original) (raw)
. 2009 Jul 2;114(1):174-80.
doi: 10.1182/blood-2009-02-207811. Epub 2009 May 7.
Sietske T Bakker, Sheba Agarwal, Michael Jansen, Elke Grassman, Barbara C Godthelp, Abdullah Mahmood Ali, Chang-hu Du, Martin A Rooimans, Qiang Fan, Kebola Wahengbam, Jurgen Steltenpool, Paul R Andreassen, David A Williams, Hans Joenje, Johan P de Winter, Amom Ruhikanta Meetei
Affiliations
- PMID: 19423727
- PMCID: PMC2710946
- DOI: 10.1182/blood-2009-02-207811
Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M
Thiyam Ramsing Singh et al. Blood. 2009.
Abstract
FANCM is a component of the Fanconi anemia (FA) core complex and one FA patient (EUFA867) with biallelic mutations in FANCM has been described. Strikingly, we found that EUFA867 also carries biallelic mutations in FANCA. After correcting the FANCA defect in EUFA867 lymphoblasts, a "clean" FA-M cell line was generated. These cells were hypersensitive to mitomycin C, but unlike cells defective in other core complex members, FANCM(-/-) cells were proficient in monoubiquitinating FANCD2 and were sensitive to the topoisomerase inhibitor camptothecin, a feature shared only with the FA subtype D1 and N. In addition, FANCM(-/-) cells were sensitive to UV light. FANCM and a C-terminal deletion mutant rescued the cross-linker sensitivity of FANCM(-/-) cells, whereas a FANCM ATPase mutant did not. Because both mutants restored the formation of FANCD2 foci, we conclude that FANCM functions in an FA core complex-dependent and -independent manner.
Figures
Figure 1
EUFA867 has biallelic FANCA mutations. (A) Introduction of FANCA in EUFA867 lymphoblasts stably expressing FANCM restores FANCD2 monoubiquitination. EUFA867 lymphoblasts were transduced with SF91-FANCM to obtain EUFA867 + FANCM. Subsequently these cells were transduced with bicistronic retroviruses encoding the indicated FA proteins. Cells were treated with 2 mM HU for 16 hours and immunoblotted for FANCD2 and FANCM. (B) EUFA867 has a nonsense mutation 2557C>T (R853X) in FANCA. (C) Mutation IVS7 + 5G>A affects the normal splice donor in FANCA intron 7 and results in a 30-bp insertion of intron 7 sequence in the cDNAs of EUFA867 and her mother. The sequence shown is from an isolated cDNA clone of EUFA867. (D) FANCA cDNA derived from the IVS7 + 5G>A mutation does not correct FANCD2 monoubiquitination. FANCA-deficient HSC72 lymphoblasts were transduced with bicistronic retrovirus encoding either wild-type FANCA or the FANCA mutant derived from the mutation IVS7 + 5G>A. Cells were treated with 1 mM HU to stimulate FANCD2 monoubiquitination.
Figure 2
Stable expression of FANCA in EUFA867 lymphoblasts partially corrects the phenotype of this cell line. (A) EUFA867 lymphoblasts stably expressing wild-type FANCA are still hypersensitive to growth inhibition by mitomycin C. FANCA-deficient HSC72 and EUFA867 lymphoblasts were transduced with SF91-FANCA. Viable cells were measured with the Cell Titer 96 Proliferation Assay. The data represent the percentage growth compared with untreated cells and show 1 representative result of 3 independent experiments with standard deviations. (B) EUFA867 lymphoblasts stably expressing wild-type FANCA show melphalan-induced G2 arrest. (C) EUFA867 lymphoblasts stably expressing wild-type FANCA have reduced FANCD2 monoubiquitination. Cells were treated with either 2 mM HU or 240 nM MMC for 16 hours or left untreated. Total lysates were immunoblotted for FANCM, FANCD2, and FANCA.
Figure 3
Ectopic expression of both FANCA and FANCM can correct the FA phenotype of EUFA867 lymphoblasts. (A) Ectopic expression of FANCA and FANCM in EUFA867 restores the chromatin localization of monoubiquitinated FANCD2, which coincides with an enhanced FANCD2 monoubiquitination. EUFA867 cells stably expressing FANCA (EUFA867-FANCA) were generated with pMMP-FANCA. Subsequently, these cells were transduced with either MIEG3 bicistronic retroviral vector or MIEG3 encoding wild-type FANCM. EGFP-positive cells were treated with 240 nM MMC for 16 hours or left untreated and subcellular fractions were made: S100 cytoplasmic and nucleoplasmic proteins, S400 chromatin-bound proteins. (B) The ATPase activity and the C-terminus of FANCM are not required for efficient FANCD2 monoubiquitination. EUFA867 lymphoblasts stably expressing FANCA were transduced with either MIEG3 retroviral vector or MIEG3 encoding wild-type FANCM, an ATPase-dead FANCM mutant (K117R-FANCM) or a C-terminal FANCM deletion mutant (delC-FANCM). EGFP-positive cells were treated with either 2 mM HU (top panel) or 240 mM MMC (bottom panel) for 16 hours. Total lysates were immunoblotted for FANCD2. (C) FANCM, but not its ATPase activity or its C-terminus, is required for the assembly of FANCD2 foci. Cells were either left untreated or exposed to 450 nM MMC or 2 mM HU for 24 hours, and the percentage of cells with 5 or more FANCD2 foci was determined in at least 150 cells. The result shows the average of 3 independent experiments with standard deviations. (D) The ATPase activity, but not the C-terminus of FANCM, is required for MMC resistance. Viable cells were measured with the Cell Titer 96 Proliferation Assay. The data represent the percentage growth compared with untreated cells and show 1 representative result of 3 independent experiments with standard deviations. (E) The FANCM ATPase mutant does not rescue the melphalan-induced G2 arrest in EUFA867 lymphoblasts. (F) FANCM is required for chromatin targeting of the FA core complex proteins. Subcellular fractions of EUFA867 lymphoblast and stably transduced derivatives were immunoblotted for FANCM, FANCA, FANCG, FANCL, and FAAP24. H2A was used as a loading control for the chromatin fraction.
Figure 4
Camptothecin sensitivity in human lymphoblasts. (A) EUFA867 lymphoblasts are sensitive to the topoisomerase I inhibitor camptothecin. Lymphoblasts were continuously exposed to different doses of camptothecin and cell growth was compared with untreated cells by cell counting. Wild-type, and FANCC- and FANCJ-deficient lymphoblasts were included as camptothecin-resistant controls. (B) Camptothecin sensitivity of EUFA867 lymphoblasts is due to a defect in FANCM. The FANCA defect in EUFA867 was corrected by stable transfection of flag-tagged FANCA (EUFA867+FANCA-flag); the FANCM defect was corrected by cell fusion with FANCA-deficient lymphoblasts HSC72 (EUFA867xHSC72OT fusion). Four independent cell fusions are depicted. (C) The C-terminus of FANCM is involved in camptothecin resistance. EUFA867 cells were stably transduced with FANCA and wild-type FANCM (FANCM) or a C-terminal deletion mutant of FANCM (delC-FANCM).
Figure 5
Camptothecin and UV sensitivity and Rad51 focus formation in human lymphoblasts. (A) FA-D1 lymphoblasts with a defect in FANCD1/BRCA2 are as sensitive to growth inhibition by camptothecin as EUFA867 lymphoblasts. (B) FA-N lymphoblasts with a defect in FANCN/PALB2 are as sensitive to growth inhibition by camptothecin as EUFA867 lymphoblasts. EUFA1341R is a MMC-resistant reverted derivative of EUFA1341 lymphoblasts. (C) EUFA867 lymphoblasts are sensitive to UV. Lymphoblasts were exposed to different doses of UVC light and cell growth was compared with untreated cells by cell counting. Lymphoblasts of xeroderma pigmentosum patient XP3BE were used as a positive control. Results show mean values of at least 5 experiments with SEM. (D) Normal Rad51 focus formation in EUFA867 lymphoblasts. Kinetics of Rad51 foci formation in response to x-ray irradiation (12 Gy) or MMC treatment (2.4 μg/mL for 1 hour) analyzed 7 and 24 hours after treatment. A cell with more than 5 distinct foci in the nucleus was considered positive. Results show the mean values of at least 2 experiments with standard error of the mean.
Similar articles
- FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair.
Xue Y, Li Y, Guo R, Ling C, Wang W. Xue Y, et al. Hum Mol Genet. 2008 Jun 1;17(11):1641-52. doi: 10.1093/hmg/ddn054. Epub 2008 Feb 19. Hum Mol Genet. 2008. PMID: 18285517 Free PMC article. - Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway.
van Twest S, Murphy VJ, Hodson C, Tan W, Swuec P, O'Rourke JJ, Heierhorst J, Crismani W, Deans AJ. van Twest S, et al. Mol Cell. 2017 Jan 19;65(2):247-259. doi: 10.1016/j.molcel.2016.11.005. Epub 2016 Dec 13. Mol Cell. 2017. PMID: 27986371 - Evidence for subcomplexes in the Fanconi anemia pathway.
Medhurst AL, Laghmani el H, Steltenpool J, Ferrer M, Fontaine C, de Groot J, Rooimans MA, Scheper RJ, Meetei AR, Wang W, Joenje H, de Winter JP. Medhurst AL, et al. Blood. 2006 Sep 15;108(6):2072-80. doi: 10.1182/blood-2005-11-008151. Epub 2006 May 23. Blood. 2006. PMID: 16720839 Free PMC article. - Molecular pathogenesis of fanconi anemia.
Taniguchi T, Dandrea AD. Taniguchi T, et al. Int J Hematol. 2002 Feb;75(2):123-8. doi: 10.1007/BF02982016. Int J Hematol. 2002. PMID: 11939257 Review. - Current knowledge on the pathophysiology of Fanconi anemia: from genes to phenotypes.
Yamashita T, Nakahata T. Yamashita T, et al. Int J Hematol. 2001 Jul;74(1):33-41. doi: 10.1007/BF02982547. Int J Hematol. 2001. PMID: 11530803 Review.
Cited by
- The differences between ICL repair during and outside of S phase.
Williams HL, Gottesman ME, Gautier J. Williams HL, et al. Trends Biochem Sci. 2013 Aug;38(8):386-93. doi: 10.1016/j.tibs.2013.05.004. Epub 2013 Jul 3. Trends Biochem Sci. 2013. PMID: 23830640 Free PMC article. Review. - FANCJ localization by mismatch repair is vital to maintain genomic integrity after UV irradiation.
Guillemette S, Branagan A, Peng M, Dhruva A, Schärer OD, Cantor SB. Guillemette S, et al. Cancer Res. 2014 Feb 1;74(3):932-44. doi: 10.1158/0008-5472.CAN-13-2474. Epub 2013 Dec 18. Cancer Res. 2014. PMID: 24351291 Free PMC article. - Inhibition of Topoisomerase (DNA) I (TOP1): DNA Damage Repair and Anticancer Therapy.
Xu Y, Her C. Xu Y, et al. Biomolecules. 2015 Jul 22;5(3):1652-70. doi: 10.3390/biom5031652. Biomolecules. 2015. PMID: 26287259 Free PMC article. Review. - The Fanconi anemia pathway: repairing the link between DNA damage and squamous cell carcinoma.
Romick-Rosendale LE, Lui VWY, Grandis JR, Wells SI. Romick-Rosendale LE, et al. Mutat Res. 2013 Mar-Apr;743-744:78-88. doi: 10.1016/j.mrfmmm.2013.01.001. Epub 2013 Jan 17. Mutat Res. 2013. PMID: 23333482 Free PMC article. Review. - Correct mRNA processing at a mutant TT splice donor in FANCC ameliorates the clinical phenotype in patients and is enhanced by delivery of suppressor U1 snRNAs.
Hartmann L, Neveling K, Borkens S, Schneider H, Freund M, Grassman E, Theiss S, Wawer A, Burdach S, Auerbach AD, Schindler D, Hanenberg H, Schaal H. Hartmann L, et al. Am J Hum Genet. 2010 Oct 8;87(4):480-93. doi: 10.1016/j.ajhg.2010.08.016. Am J Hum Genet. 2010. PMID: 20869034 Free PMC article.
References
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. - PubMed
- Meetei A, de Winter J, Medhurst A, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. - PubMed
- Machida YJ, Machida Y, Chen Y, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous