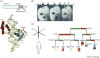Regeneration, repair and remembering identity: the three Rs of Hox gene expression - PubMed (original) (raw)
Review
Regeneration, repair and remembering identity: the three Rs of Hox gene expression
Kevin C Wang et al. Trends Cell Biol. 2009 Jun.
Abstract
Hox genes encode transcription factors that specify embryonic positional identity in cells and guide tissue differentiation. Recent advances have greatly increased our understanding of the epigenetic mechanisms that ensure the faithful expression of Hox genes in adult cells and which involve the interplay of histone methylation, demethylation and intergenic transcription of long non-coding RNAs. The transcriptional memory of Hox genes poses both an opportunity and a challenge for regenerative medicine. Matching the positional identity of transplanted stem cells with that of the host environment, as reflected by their respective Hox profiles, is likely to be required to achieve regenerative healing. Strategies to manipulate the plasticity of Hox gene expression will probably become a major focus in regenerative medicine.
Figures
Figure 1
Hox genes and positional identity. (a) Hox genes belong in the family of homeodomain-containing transcription factors. The domain organization of HOXA5 and the crystal structure of the homologous fly Scr Hox protein (brown) in complex with Exd (light blue), binding to DNA (green) are shown. Exd is a Hox cofactor that acts as an accessory DNA-binding factor. Homeodomains are helix-turn-helix motifs (represented by α1, α2 and α3) that bind specific DNA sequences. Sequences N-terminal of the homeodomain, such as the YPWM motif, are involved in protein–protein interaction with accessory DNA-binding factors such as Exd. Part (a) adapted, with permission, from Ref. [44]. (b) Expression of three HoxD genes in developing mouse embryos by in situ hybridization. The schematic above the in situ pictures represents the HoxD locus – the individual numbers refer to different HoxD genes. The white arrows indicate that HoxD-13, -11 and -9 are shown in the three panels, from left to right. The orange and blue dotted lines represent the neural tube and paraxial mesoderm, respectively – the lines are there to highlight the tissue-specific mechanisms for colinear gene regulation: Hox genes tend to have a very distinctive organization in the genome, being arranged in gene clusters in which the order of the genes within the cluster corresponds to (or is ‘colinear with’) some aspect of the gene expression. In addition to spatial colinearity, in vertebrates there is also a temporal colinearity, where the 3′–5′ arrangement of genes also reflects the temporal order in which they are activated during development. The expression domains of Hox genes towards the 3′ UTR seem closer to the head and closer to the trunk (dotted lines), whereas the Hox genes towards the 5′ UTR are expressed closer to the tail and closer to fingers or toes on the limbs (pins). For example, the domains of expression of _HoxD_13 in both the neural tube (marker by the orange line) and the paraxial mesoderm (the blue line) are quite similar, whereas in _HoxD_11 and to a greater extent _HoxD_9, one can really appreciate the variation in the transcript domains of the same gene in different tissues. The pins represent nested domains of expression, from the highest level (black) to lowest (white), again illustrating the colinearity principle. Part (b) reproduced, with permission, from Ref. [45]. (c) HOX genes as the address code of the human body. As illustrated by the decision tree, differential expression of HOX genes reflects the anatomic origin of adult human fibroblasts along the anterior (A)–posterior (P), dorso (D)–ventral (V), and proximal (Px)–distal (Ds) axes, in addition to their origin from cutaneous versus internal organs. Red indicates high expression; green indicates low expression.
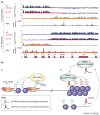
Figure 2
Epigenetic landscape of Hox genes. (a) Chromatin state map across~100 kilobases of the human HOXA locus (X axis) obtained from chromatin IP on microarray (ChIP-chip) experiments, a technique for isolation and identification of the DNA sequences occupied by specific DNA-binding proteins in cells. Y axes indicate the occupancy of the indicated proteins (represented by the log2 ratio of ChIP over input) or expression of RNAs (represented by a linear scale of hybridization intensity). Suz12 is a component of the PRC2 that mediates histone H3 Lys27 methylation (H3K27me3). RNA polymerase (Pol) II broadly occupies the regions that are also transcribed and occupied by H3K4me3 (not shown). Note that fibroblasts from two different anatomic origins (lung and foot) can program the HOXA locus in diametrically opposite ways but along the same boundary. The HOXA locus is depicted at the bottom of the map, with HoxA13 at the distal 5′ end and HoxA1 at the promixal 3′ end. The dashed line highlights the boundary of opposite configurations of chromatin modification and intergenic transcription. (b) Model of chromatin state regulation by lncRNAs. lncRNAs can affect chromatin state in cis or in trans, and in gene activation or silencing, via histone-modification enzymes. (i) lncRNAs might increase the accessibility of Trithorax group proteins such as ASH1 (absent, small, or homeotic discs 1) or MLL or directly recruit them, leading to trimethylation of histone H3K4 and transcriptional activation of downstream targets such as the Hox genes. (ii) By contrast, lncRNAs might also target the PRC2 proteins to trimethylate H3K27 at a distance and render the target genes transcriptionally silent.
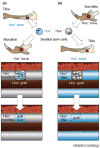
Figure 3
Hox status and bone regeneration in mouse. Cells expressing Hox genes (for example Hoxa11) are in light blue and those not expressing Hox are indicated in light grey. Hox+ skeletal stem cells can only heal orthotopic Hox+ bone injury site (tibia). (a) Hox+ skeletal stem cells cannot repair a _Hox_− injury site (mandible). (b) Conversely, _Hox_− skeletal stem cells will express the ectopic Hox gene when transplanted into a Hox+ injury site and regenerate the ectopic bone.
Similar articles
- Hox genes in the adult skeleton: Novel functions beyond embryonic development.
Rux DR, Wellik DM. Rux DR, et al. Dev Dyn. 2017 Apr;246(4):310-317. doi: 10.1002/dvdy.24482. Epub 2017 Jan 27. Dev Dyn. 2017. PMID: 28026082 Free PMC article. Review. - [Hox Genes and Animal Regeneration].
Novikova EL, Bakalenko NI, Nesterenko AY, Kulakova MA. Novikova EL, et al. Ontogenez. 2016 Jul-Aug;47(4):209-18. Ontogenez. 2016. PMID: 30272395 Review. Russian. - Epigenetic regulations in hematopoietic Hox code.
He H, Hua X, Yan J. He H, et al. Oncogene. 2011 Jan 27;30(4):379-88. doi: 10.1038/onc.2010.484. Epub 2010 Oct 25. Oncogene. 2011. PMID: 20972460 Review. - HOX and TALE signatures specify human stromal stem cell populations from different sources.
Picchi J, Trombi L, Spugnesi L, Barachini S, Maroni G, Brodano GB, Boriani S, Valtieri M, Petrini M, Magli MC. Picchi J, et al. J Cell Physiol. 2013 Apr;228(4):879-89. doi: 10.1002/jcp.24239. J Cell Physiol. 2013. PMID: 23018864 - Homeobox gene expression profile in human hematopoietic multipotent stem cells and T-cell progenitors: implications for human T-cell development.
Taghon T, Thys K, De Smedt M, Weerkamp F, Staal FJ, Plum J, Leclercq G. Taghon T, et al. Leukemia. 2003 Jun;17(6):1157-63. doi: 10.1038/sj.leu.2402947. Leukemia. 2003. PMID: 12764384
Cited by
- Potential role of the HOXD8 transcription factor in cisplatin resistance and tumour metastasis in advanced epithelial ovarian cancer.
Sun P, Song Y, Liu D, Liu G, Mao X, Dong B, Braicu EI, Sehouli J. Sun P, et al. Sci Rep. 2018 Sep 7;8(1):13483. doi: 10.1038/s41598-018-31030-3. Sci Rep. 2018. PMID: 30194340 Free PMC article. - Impact of HOXB7 overexpression on human adipose-derived mesenchymal progenitors.
Foppiani EM, Candini O, Mastrolia I, Murgia A, Grisendi G, Samarelli AV, Boscaini G, Pacchioni L, Pinelli M, De Santis G, Horwitz EM, Veronesi E, Dominici M. Foppiani EM, et al. Stem Cell Res Ther. 2019 Mar 19;10(1):101. doi: 10.1186/s13287-019-1200-6. Stem Cell Res Ther. 2019. PMID: 30890185 Free PMC article. - Locational memory of macrovessel vascular cells is transcriptionally imprinted.
Spanjersberg TCF, Oosterhoff LA, Kruitwagen HS, van den Dungen NAM, Vernooij JCM, Asselbergs FW, Mokry M, Spee B, Harakalova M, van Steenbeek FG. Spanjersberg TCF, et al. Sci Rep. 2023 Aug 10;13(1):13028. doi: 10.1038/s41598-023-38880-6. Sci Rep. 2023. PMID: 37563195 Free PMC article. - A combination analysis based on bioinformatics tools reveals new signature genes related to maternal obesity and fetal programming.
Liu C, Lu Y, Huang C, Zeng Y, Zheng Y, Wang C, Huang H. Liu C, et al. Front Med (Lausanne). 2024 Sep 4;11:1434105. doi: 10.3389/fmed.2024.1434105. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39296904 Free PMC article. - Recurrent oxidant treatment induces dysregulation in the brain transcriptome of Atlantic salmon (Salmo salar) smolts.
Carletto D, Breiland MW, Hytterød S, Timmerhaus G, Lazado CC. Carletto D, et al. Toxicol Rep. 2022 Jun 23;9:1461-1471. doi: 10.1016/j.toxrep.2022.06.009. eCollection 2022. Toxicol Rep. 2022. PMID: 36518465 Free PMC article.
References
- McGonigle GJ, et al. Grappling with the HOX network in hematopoiesis and leukemia. Front Biosci. 2008;13:4297–4308. - PubMed
- Maroulakou IG, Spyropoulos DD. The study of HOX gene function in hematopoietic, breast and lung carcinogenesis. Anticancer Res. 2003;23:2101–2110. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources