Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux - PubMed (original) (raw)
Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux
Tomohiro Shimada et al. J Bacteriol. 2009 Jul.
Abstract
LeuO, a LysR family transcription factor, exists in a wide variety of bacteria of the family Enterobacteriaceae and is involved in the regulation of as yet unidentified genes affecting the stress response and pathogenesis expression. Using genomic screening by systematic evolution of ligands by exponential enrichment (SELEX) in vitro, a total of 106 DNA sequences were isolated from 12 different regions of the Escherichia coli genome. All of the SELEX fragments formed complexes in vitro with purified LeuO. After Northern blot analysis of the putative target genes located downstream of the respective LeuO-binding sequence, a total of nine genes were found to be activated by LeuO, while three genes were repressed by LeuO. The LeuO target gene collection included several multidrug resistance genes. A phenotype microarray assay was conducted to identify the gene(s) responsible for drug resistance and the drug species that are under the control of the LeuO target gene(s). The results described herein indicate that the yjcRQP operon, one of the LeuO targets, is involved in sensitivity control against sulfa drugs. We propose to rename the yjcRQP genes the sdsRQP genes (sulfa drug sensitivity determinant).
Figures
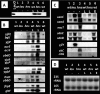
FIG. 1.
Gene organization of operons that form complexes in vitro with purified LeuO protein. (A) Operons that are activated in vivo by LeuO, as detected by Northen blot analysis (see Fig. 3). (B) Operons that are repressed by LeuO. Symbols: closed circles, promoters; open squares, H-NS binding sites (from the work of Oshima et al. [41]).
FIG. 2.
Gel shift assay of LeuO-SELEX fragment interaction. An 8 nM concentration of each of the indicated fluorescently labeled DNA probes was incubated at 37°C for 15 min in the absence (lanes 1) or presence of 2.0 (lanes 2), 4.0 (lanes 3), and 8.0 nM (lanes 4) purified LeuO and then directly subjected to electrophoresis on a 5% polyacrylamide gel.
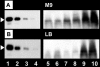
FIG. 3.
Intracellular level of LeuO protein. Wild-type E. coli was grown at 37°C in either M9-0.4% glucose medium (A) or LB medium (B) for various times and then subjected to quantitative Western blot analysis (26) for determination of the LeuO level. Lanes 1 to 4, 10, 5, 2.5, and 1 ng purified LeuO protein; lanes 5 to 10, extracts of cells harvested at OD600s of 0.15, 0.3, 0.6, 1.2, 5.0 (48 h), and 5.0 (72 h).
FIG. 4.
Northern blot analysis of LeuO-dependent gene transcripts. Wild-type E. coli BW25113 (lanes 1; wt), leuO deletion mutant JW0075 (lanes 2; leu), hns deletion mutant JW1225 (lanes 3; hns), the wild type with pLeuO (lanes 4; wt), the hns mutant with pLeuO (lanes 5; hns), and the wild type with empty vector (lanes 6; wt) were grown at 37°C in LB medium. (A) Whole-cell extracts were prepared from cells harvested at an OD600 of 0.5 and subjected to Western blot analysis against anti-LeuO protein. (B) Total RNA was isolated from each culture, and 4 μg was fractionated by PAGE in the presence of urea. After blotting of RNAs onto filters, transcripts of the LeuO target genes were detected with the fluorescently labeled probes indicated on the left. (C) Total RNA was subjected to Northern blot analysis as in panel B. Probes used are indicated on the left. (D) Total RNAs used for Northern blot analysis were fractionated by PAGE, and the gel was stained with ethidium bromide for detection of 16S and 23S rRNAs.
FIG. 5.
Sulfadiazine sensitivity test with mutants lacking the multidrug pump genes. E. coli BW25113 (WT), the JW0451 acrE mutant (acrE), the JW2730 ygcL mutant (ygcL), and the JW4041 mdtN mutant (yjcR), each with and without the LeuO expression plasmid, were grown in the presence of increasing concentrations of sulfadiazine for 8 h. The concentrations of sulfadiazine were as follows (from left to right): 10 μg/ml in the first column, followed by twofold dilutions to the 11th column; the last column contained no drug. Cell growth was monitored by measuring respiration (tetrazolium color formation).
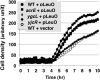
FIG. 6.
Effect of sulfadiazine on growth. E. coli BW25113 (WT), the JW0451 acrE mutant (acrE), the JW2730 ygcL mutant (ygcL), and the JW4041 mdtN mutant (yjcR), each carrying either the LeuO expression plasmid pLeuO or empty plasmid, were grown in the presence or absence of 5 μg/ml sulfadiazine. Cell growth was monitored by measuring respiration (tetrazolium color formation).
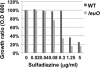
FIG. 7.
Sensitivities of wild-type and leuO mutant E. coli cells to sulfamidazine. Overnight cultures of wild-type and leuO mutant E. coli in M9-0.4% glucose medium at 37°C were transferred into fresh M9-0.4% glucose medium containing the indicated concentrations of sulfamidazine. Growth was monitored by measuring the turbidity (OD600). After 12 h of culture, the relative rates of cell growth in the presence and absence of drug were plotted.
Similar articles
- Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS.
Shimada T, Bridier A, Briandet R, Ishihama A. Shimada T, et al. Mol Microbiol. 2011 Oct;82(2):378-97. doi: 10.1111/j.1365-2958.2011.07818.x. Epub 2011 Sep 14. Mol Microbiol. 2011. PMID: 21883529 - Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO.
Stratmann T, Madhusudan S, Schnetz K. Stratmann T, et al. J Bacteriol. 2008 Feb;190(3):926-35. doi: 10.1128/JB.01447-07. Epub 2007 Nov 30. J Bacteriol. 2008. PMID: 18055596 Free PMC article. - Activation of leuO by LrhA in Escherichia coli.
Breddermann H, Schnetz K. Breddermann H, et al. Mol Microbiol. 2017 May;104(4):664-676. doi: 10.1111/mmi.13656. Epub 2017 Mar 22. Mol Microbiol. 2017. PMID: 28252809 - The LeuO regulator and quiescence: About transcriptional roadblocks, multiple promoters, and crispr-cas.
Sánchez-Popoca D, Serrano-Fujarte I, Fernández-Mora M, Calva E. Sánchez-Popoca D, et al. Mol Microbiol. 2022 Nov;118(5):503-509. doi: 10.1111/mmi.14990. Epub 2022 Oct 20. Mol Microbiol. 2022. PMID: 36203248 Review. - Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella.
Nishino K, Nikaido E, Yamaguchi A. Nishino K, et al. Biochim Biophys Acta. 2009 May;1794(5):834-43. doi: 10.1016/j.bbapap.2009.02.002. Epub 2009 Feb 20. Biochim Biophys Acta. 2009. PMID: 19230852 Review.
Cited by
- The Subtleties and Contrasts of the LeuO Regulator in Salmonella Typhi: Implications in the Immune Response.
Guadarrama C, Villaseñor T, Calva E. Guadarrama C, et al. Front Immunol. 2014 Dec 12;5:581. doi: 10.3389/fimmu.2014.00581. eCollection 2014. Front Immunol. 2014. PMID: 25566242 Free PMC article. Review. - BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS.
Venkatesh GR, Kembou Koungni FC, Paukner A, Stratmann T, Blissenbach B, Schnetz K. Venkatesh GR, et al. J Bacteriol. 2010 Dec;192(24):6456-64. doi: 10.1128/JB.00807-10. Epub 2010 Oct 15. J Bacteriol. 2010. PMID: 20952573 Free PMC article. - The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi.
Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. Medina-Aparicio L, et al. J Bacteriol. 2011 May;193(10):2396-407. doi: 10.1128/JB.01480-10. Epub 2011 Mar 11. J Bacteriol. 2011. PMID: 21398529 Free PMC article. - NhaR, LeuO, and H-NS Are Part of an Expanded Regulatory Network for Ectoine Biosynthesis Expression.
Boas Lichty KE, Gregory GJ, Boyd EF. Boas Lichty KE, et al. Appl Environ Microbiol. 2023 Jun 28;89(6):e0047923. doi: 10.1128/aem.00479-23. Epub 2023 Jun 6. Appl Environ Microbiol. 2023. PMID: 37278653 Free PMC article. - A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions.
Teramoto J, Yoshimura SH, Takeyasu K, Ishihama A. Teramoto J, et al. Nucleic Acids Res. 2010 Jun;38(11):3605-18. doi: 10.1093/nar/gkq077. Epub 2010 Feb 15. Nucleic Acids Res. 2010. PMID: 20156994 Free PMC article.
References
- Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 25611905-11910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials