Chromosome segregation machinery and cancer - PubMed (original) (raw)
Review
Chromosome segregation machinery and cancer
Kozo Tanaka et al. Cancer Sci. 2009 Jul.
Abstract
Loss or gain of chromosomes is associated with many cancer cells. This property, called chromosome instability, might arise from a lesion in the chromosome segregation machinery. Essential for chromosome segregation are the proper connection of microtubules to kinetochores, and the synchronous segregation of sister chromatids in anaphase. Accuracy of these processes is ensured by two sophisticated machineries called the correction mechanism and the spindle assembly checkpoint. Here we outline the current understanding of the underlying mechanisms, and highlight recent challenging experiments to address how chromosome segregation failure might relate to tumorigenesis. Understanding these mechanisms may lead to the discovery of new and improved anticancer therapies.
Figures
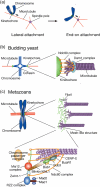
Figure 1
Kinetochore–microtubule attachment. (a) Lateral versus end‐on attachment. Kinetochores are initially captured by the lattice of microtubules (lateral attachment; left) and are subsequently tethered at the end of microtubules (end‐on attachment; right). In both cases, kinetochores are transported toward a spindle pole (arrows). (b) Kinetochore–microtubule attachment in budding yeast. The Ndc80 complex is at the interface of the kinetochore–microtubule interaction, whereas the Dam1 complex is involved in poleward kinetochore motion during end‐on attachment. (c) Kinetochore–microtubule attachment in metazoans. Correlation between the structures found in electron microscopy (upper right panel) and the kinetochore structure defined at the molecular level (lower panel) has not been precisely determined yet. Molecules involved in the chromosome segregation machinery are shown schematically. Note that spindle assembly checkpoint components (Mad1, Mad2, Bub1, BubR1, and Bub3) are delocalized from kinetochores when proper kinetochore–microtubule attachment is established. Bub, budding uninhibited by benzimidazole; BubR1, Bub1‐related 1; CENP‐E, centromere protein E; Dam, Duo1 and Mps1‐interacting; KNL, kinetochore‐null; Mad, mitotic arrest‐deficient; Mis, minichromosome instability; Ndc, nuclear division cycle; RZZ; Rod, ZW10, and ZWILCH; Zwint ZW10 interactor.
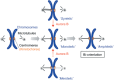
Figure 2
How chromosomes bi‐orient on the mitotic spindle. As the capture of kinetochores by microtubules is a stochastic process, wrong orientations, termed syntelic or merotelic attachments, can occur. Because the correction mechanism is mediated by Aurora B, inhibition of this kinase results in an increased frequency of these erroneous attachments.
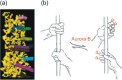
Figure 3
Interaction of microtubule plus‐ends with the kinetochore outer plate. (a) The fibrous network structure of the kinetochore outer plate, as revealed by electron tomography.( 10 ) The microtubule ends appear to be embedded in this fibrous network, and some of the fibers extend out from the plate and physically contact the microtubule wall. (Adapted by permission from Macmillan Publishers: McEwen et al. 2007 Nat. Cell Biol. 9: 516–522, copyright 2007.) (b) A cartoon illustrating how dynamic binding of kinetochores and microtubules are regulated. The affinity between fibrous structures of the outer plate (denoted by hands), and the microtubule (pole), is controlled by Aurora B‐mediated phosphorylation of the outer‐plate fibers.
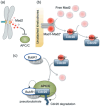
Figure 4
Molecular basis of the SAC. (a) The SAC ‘wait anaphase’ signal is generated at unattached kinetochores. By contacting the unattached kinetochores, Mad2 molecules become active (denoted by a color change from light to dark red), eventually leading to APC/C inhibition. (b) Two different conformations of Mad2 are shown in dark and light red circles. Most of the cytoplamic, free Mad2 (light red) is converted to an alternative conformer that can bind Cdc20 (dark red). The template model predicts that the Mad1–Mad2 complex at unattached kinetochores as well as the Cdc20–Mad2 complex can both catalyze this conformational change. (c) Cdc20 is handed over from Mad2 to the Bub1‐related 1 BubR1‐Bub3 complex. BubR1 inhibits APC/C activity by acting as a psuedosubstrate, and/or by mediating Cdc20 ubiquitination and degradation (as denoted by the dotted arrows).( 48 ) It is not entirely clear when the APC/C recruits Cdc20 in this cascade.
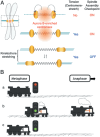
Figure 5
Kinetochore stretching inactivates the SAC. (A) SAC inactivation depends on kinetochore stretching rather than the tension‐induced stretch of centromeres between sister kinetochores. When tension is low, the microtubule attachment sites (i.e. kinetochores) position close to Aurora B‐enriched centromeres where microtubule‐releasing activity is thought to be high. (B) A cartoon modeling how the metaphase‐to‐anaphase transition is controlled. (a) As a locomotive travels from metaphase to anaphase, the presence of unattached kinetochores sustains the canonical SAC pathway and generates a strong ‘wait anaphase’ signal (red light). (b) After microtubule attachment is fulfilled (green light), kinetochore stretching (denoted by a pump inside the engine) facilitates inactivation of SAC (i.e. activation of the anaphase‐promoting complex or cyclosome) throughout the metaphase‐to‐anaphase transition. (c) But if kinetochore stretching is perturbed the locomotive does not move forward because there is no pumping in the engine.
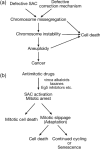
Figure 6
Mitotic regulation and cancer. (a) A model for how mitotic dysregulation could promote tumorigenesis. (b) Possible outcomes for the cell following treatment with antimitotic drugs. SAC, spindle assembly checkpoint.
Similar articles
- Attachment and tension in the spindle assembly checkpoint.
Zhou J, Yao J, Joshi HC. Zhou J, et al. J Cell Sci. 2002 Sep 15;115(Pt 18):3547-55. doi: 10.1242/jcs.00029. J Cell Sci. 2002. PMID: 12186941 Review. - Monitoring the fidelity of mitotic chromosome segregation by the spindle assembly checkpoint.
Silva P, Barbosa J, Nascimento AV, Faria J, Reis R, Bousbaa H. Silva P, et al. Cell Prolif. 2011 Oct;44(5):391-400. doi: 10.1111/j.1365-2184.2011.00767.x. Cell Prolif. 2011. PMID: 21951282 Free PMC article. Review. - Merotelic kinetochores in mammalian tissue cells.
Salmon ED, Cimini D, Cameron LA, DeLuca JG. Salmon ED, et al. Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):553-68. doi: 10.1098/rstb.2004.1610. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897180 Free PMC article. Review. - Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes.
Cimini D, Cameron LA, Salmon ED. Cimini D, et al. Curr Biol. 2004 Dec 14;14(23):2149-55. doi: 10.1016/j.cub.2004.11.029. Curr Biol. 2004. PMID: 15589159 - Chromosome segregation: correcting improper attachment.
Kapoor TM. Kapoor TM. Curr Biol. 2004 Dec 14;14(23):R1011-3. doi: 10.1016/j.cub.2004.11.026. Curr Biol. 2004. PMID: 15589138 Review.
Cited by
- Chromatin Separation Regulators Predict the Prognosis and Immune Microenvironment Estimation in Lung Adenocarcinoma.
Li Z, Ma Z, Xue H, Shen R, Qin K, Zhang Y, Zheng X, Zhang G. Li Z, et al. Front Genet. 2022 Jul 8;13:917150. doi: 10.3389/fgene.2022.917150. eCollection 2022. Front Genet. 2022. PMID: 35873497 Free PMC article. - Essential role for centromeric factors following p53 loss and oncogenic transformation.
Filipescu D, Naughtin M, Podsypanina K, Lejour V, Wilson L, Gurard-Levin ZA, Orsi GA, Simeonova I, Toufektchan E, Attardi LD, Toledo F, Almouzni G. Filipescu D, et al. Genes Dev. 2017 Mar 1;31(5):463-480. doi: 10.1101/gad.290924.116. Epub 2017 Mar 29. Genes Dev. 2017. PMID: 28356341 Free PMC article. - Kdm4c is Recruited to Mitotic Chromosomes and Is Relevant for Chromosomal Stability, Cell Migration and Invasion of Triple Negative Breast Cancer Cells.
Garcia J, Lizcano F. Garcia J, et al. Breast Cancer (Auckl). 2018 Jul 27;12:1178223418773075. doi: 10.1177/1178223418773075. eCollection 2018. Breast Cancer (Auckl). 2018. PMID: 30083054 Free PMC article. - CAMP (C13orf8, ZNF828) is a novel regulator of kinetochore-microtubule attachment.
Itoh G, Kanno S, Uchida KS, Chiba S, Sugino S, Watanabe K, Mizuno K, Yasui A, Hirota T, Tanaka K. Itoh G, et al. EMBO J. 2011 Jan 5;30(1):130-44. doi: 10.1038/emboj.2010.276. Epub 2010 Nov 9. EMBO J. 2011. PMID: 21063390 Free PMC article. - Knockdown of dystrophin Dp71 impairs PC12 cells cycle: localization in the spindle and cytokinesis structures implies a role for Dp71 in cell division.
Villarreal-Silva M, Centeno-Cruz F, Suárez-Sánchez R, Garrido E, Cisneros B. Villarreal-Silva M, et al. PLoS One. 2011;6(8):e23504. doi: 10.1371/journal.pone.0023504. Epub 2011 Aug 19. PLoS One. 2011. PMID: 21886794 Free PMC article.
References
- Tanaka TU. Bi‐orienting chromosomes: acrobatics on the mitotic spindle. Chromosoma 2008; 117: 521–33. - PubMed
- Tanaka TU, Stark MJ, Tanaka K. Kinetochore capture and bi‐orientation on the mitotic spindle. Nat Rev Mol Cell Biol 2005; 6: 929–42. - PubMed
- Cheeseman IM, Chappie JS, Wilson‐Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule‐binding site of the kinetochore. Cell 2006; 127: 983–97. - PubMed
- Tanaka K, Mukae N, Dewar H et al . Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 2005; 434: 987–94. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources