ace, Which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence - PubMed (original) (raw)
ace, Which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence
Francois Lebreton et al. Infect Immun. 2009 Jul.
Abstract
Enterococcus faecalis is an opportunistic pathogen that causes numerous infectious diseases in humans and is a major agent of nosocomial infections. In this work, we showed that the recently identified transcriptional regulator Ers (PrfA like), known to be involved in the cellular metabolism and the virulence of E. faecalis, acts as a repressor of ace, which encodes a collagen-binding protein. We characterized the promoter region of ace, and transcriptional analysis by reverse transcription-quantitative PCR and mobility shift protein-DNA binding assays revealed that Ers directly regulates the expression of ace. Transcription of ace appeared to be induced by the presence of bile salts, probably via the deregulation of ers. Moreover, with an ace deletion mutant and the complemented strain and by using an insect (Galleria mellonella) virulence model, as well as in vivo-in vitro murine macrophage models, we demonstrated for the first time that Ace can be considered a virulence factor for E. faecalis. Furthermore, animal experiments revealed that Ace is also involved in urinary tract infection by E. faecalis.
Figures
FIG. 1.
(A) Sequence of the ace promoter region. Potential −35 and −10 region and ribosome binding site (RBS) sequences are underlined. The transcriptional start site (+1) is in boldface, and the Ers box is boxed. (B) EMSA with the promoter region of the ace gene and different concentrations (2 to 0.2 μg) of His6-ErsHTH protein. (C) EMSA of His6-ErsHTH binding to the D4-labeled DNA fragment containing the ace regulatory region. Amplifications were performed with D4-Pu and D4-Pr (Table 2). Crude cell extracts prepared from an E. faecalis Δ_ers_ mutant strain (0.1 μg protein) were added to all of the reaction mixtures. Shown are the labeled ace promoter without protein (lane 1), with His6-ErsHTH (1 μg protein) (lane 2), with His6-ErsHTH and an unlabeled competitor (lane 3), and with His6-ErsHTH and a nonspecific DNA fragment (internal fragment of the ef1843 gene) (lane 4).
FIG. 2.
Expression of ers and ace determined by Western blot or RT-qPCR analysis. (A) Western blot analysis (with antiserum against His-tagged Ers) of E. faecalis protein extracts (40 μg) from strain JH2-2 grown at 37°C (lane 2), at 46°C (lane 3), or in the presence of bile salts (lane 4) and from strain JH2-2/p3535_ers_ (lane 5). A 0.1-μg sample of purified His6-Ers protein was loaded into lane 1. (B) RT-qPCRs experiments with the ers and ace genes and primers described in Table 2. RNAs were extracted from cells cultured under the same conditions used for protein extracts. Values that are significantly different are >2 (P < 0.05).
FIG. 3.
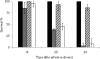
Effect of ace inactivation on virulence. Percent survival of G. mellonella larvae at 16, 20, and 24 h after infection with L. lactis IL1403 (black bar), E. faecalis JH2-2 (gray bar), the JH2-2 Δ_ace_ mutant (hatched bar), and the complemented Δ_ace_ mutant strain (white bar). We used 6 × 106 CFU counted on an agar plate per injection. Experiments were repeated at least three times, and the results represent the mean ± standard deviation of live larvae.
FIG. 4.
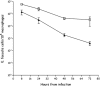
Time course of intracellular survival of E. faecalis JH2-2 (squares) and the Δ_ace_ mutant (triangles) within murine peritoneal macrophages. The results shown represent the mean number ± the standard deviation of viable intracellular bacteria per 105 macrophages of three independent experiments with three wells.
FIG. 5.
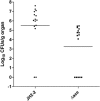
Infection with 104 cells of wild-type E. faecalis JH2-2 (•) and its isogenic Δ_ace_ mutant strain (▪). Kidney pair homogenates were obtained from groups of 15 mice that were sacrificed and necropsied 48 h after a transurethral challenge. Results are expressed as log10 CFU per gram of tissue. A value of 0 was assigned to uninfected kidneys. Horizontal bars represent the geometric means.
Similar articles
- The two-component system GrvRS (EtaRS) regulates ace expression in Enterococcus faecalis OG1RF.
Roh JH, Singh KV, La Rosa SL, Cohen AL, Murray BE. Roh JH, et al. Infect Immun. 2015 Jan;83(1):389-95. doi: 10.1128/IAI.02587-14. Epub 2014 Nov 10. Infect Immun. 2015. PMID: 25385790 Free PMC article. - Expression of the collagen adhesin ace by Enterococcus faecalis strain OG1RF is not repressed by Ers but requires the Ers box.
Cohen AL, Roh JH, Nallapareddy SR, Höök M, Murray BE. Cohen AL, et al. FEMS Microbiol Lett. 2013 Jul;344(1):18-24. doi: 10.1111/1574-6968.12146. Epub 2013 May 1. FEMS Microbiol Lett. 2013. PMID: 23551253 Free PMC article. - Expression of Adhesive Pili and the Collagen-Binding Adhesin Ace Is Activated by ArgR Family Transcription Factors in Enterococcus faecalis.
Manias DA, Dunny GM. Manias DA, et al. J Bacteriol. 2018 Aug 24;200(18):e00269-18. doi: 10.1128/JB.00269-18. Print 2018 Sep 15. J Bacteriol. 2018. PMID: 29986940 Free PMC article. - Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches.
Ali L, Goraya MU, Arafat Y, Ajmal M, Chen JL, Yu D. Ali L, et al. Int J Mol Sci. 2017 May 3;18(5):960. doi: 10.3390/ijms18050960. Int J Mol Sci. 2017. PMID: 28467378 Free PMC article. Review. - Lipoproteins of Enterococcus faecalis: bioinformatic identification, expression analysis and relation to virulence.
Reffuveille F, Leneveu C, Chevalier S, Auffray Y, Rincé A. Reffuveille F, et al. Microbiology (Reading). 2011 Nov;157(Pt 11):3001-3013. doi: 10.1099/mic.0.053314-0. Epub 2011 Sep 8. Microbiology (Reading). 2011. PMID: 21903750 Review.
Cited by
- Architectural dissection of adhesive bacterial cell surface appendages from a "molecular machines" viewpoint.
Smith OER, Bharat TAM. Smith OER, et al. J Bacteriol. 2024 Nov 5:e0029024. doi: 10.1128/jb.00290-24. Online ahead of print. J Bacteriol. 2024. PMID: 39499080 Free PMC article. Review. - Enterococcal-host interactions in the gastrointestinal tract and beyond.
Madani WAM, Ramos Y, Cubillos-Ruiz JR, Morales DK. Madani WAM, et al. FEMS Microbes. 2024 Sep 9;5:xtae027. doi: 10.1093/femsmc/xtae027. eCollection 2024. FEMS Microbes. 2024. PMID: 39391373 Free PMC article. Review. - Enterococcus faecalis: an overlooked cell invader.
Archambaud C, Nunez N, da Silva RAG, Kline KA, Serror P. Archambaud C, et al. Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0006924. doi: 10.1128/mmbr.00069-24. Epub 2024 Sep 6. Microbiol Mol Biol Rev. 2024. PMID: 39239986 Review. - Detecting co-selection through excess linkage disequilibrium in bacterial genomes.
Mallawaarachchi S, Tonkin-Hill G, Pöntinen AK, Calland JK, Gladstone RA, Arredondo-Alonso S, MacAlasdair N, Thorpe HA, Top J, Sheppard SK, Balding D, Croucher NJ, Corander J. Mallawaarachchi S, et al. NAR Genom Bioinform. 2024 Jun 6;6(2):lqae061. doi: 10.1093/nargab/lqae061. eCollection 2024 Jun. NAR Genom Bioinform. 2024. PMID: 38846349 Free PMC article. - Promiscuous, persistent and problematic: insights into current enterococcal genomics to guide therapeutic strategy.
Hourigan D, Stefanovic E, Hill C, Ross RP. Hourigan D, et al. BMC Microbiol. 2024 Mar 28;24(1):103. doi: 10.1186/s12866-024-03243-2. BMC Microbiol. 2024. PMID: 38539119 Free PMC article. Review.
References
- Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44183-190. - PubMed
- Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11260-263. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous