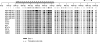The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer - PubMed (original) (raw)
The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer
R Shah et al. Br J Cancer. 2009.
Abstract
Expression of P3H2 (Leprel1) and P3H3 (Leprel2) but not P3H1 (Leprecan) is down-regulated in breast cancer by aberrant CpG methylation in the 5' regulatory sequences of each gene. Methylation of P3H2 appears specific to breast cancer as no methylation was detected in a range of cell lines from other epithelial cancers or from primary brain tumours or malignant melanoma. Methylation in P3H2, but not P3H3, was strongly associated with oestrogen-receptor-positive breast cancers, whereas methylation in P3H3 was associated with higher tumour grade and Nottingham Prognostic Index. Ectopic expression of P3H2 and P3H3 in cell lines with silencing of the endogenous gene results in suppression of colony growth. This is the first demonstration of epigenetic inactivation of prolyl hydroxylases in human cancer, implying that this gene family represents a novel class of tumour suppressors. The restriction of silencing in P3H2 to breast carcinomas, and its association with oestrogen-receptor-positive cases, suggests that P3H2 may be a breast-cancer-specific tumour suppressor.
Figures
Figure 1
Epigenetic regulation of expression of P3H2 and P3H3 in breast carcinoma cell lines. (A) RT–PCR analysis of P3H1, P3H2 and P3H3 expression in the indicated breast carcinoma cell lines and normal breast epithelium (HMEC). The control gene GAPDH is also shown. (B) Western blot analysis of expression of P3H2 and P3H3 in breast carcinoma cell lines. Western blot analysis of the indicated breast carcinoma cell lines was performed as described in Materials and methods. The control gene PCNA is also shown. Approximate position of molecular weight markers is indicated. (C) Methylation in the CpG islands of P3H2 and P3H3 correlates with down-regulation of expression. As shown, the CpG island of P3H1 was uniformly unmethylated in each cell line and in normal breast epithelium (HMEC), consistent with expression analysis (A). The figure shows MSP analysis of the P3H2 CpG island using two primer pairs. Pair 1 (upper panel of P3H2) detects methylation in the MDA MB 453 and T47D cell lines, both of which lacked detectable expression. Primer pair 2 (lower panel of P3H2) that is located further 3′ in the CpG island detected methylation in MDA MB 361, MDA MB 453, MCF7, T47D, BT474 and SKBR3 cell lines, which correlates closely with expression analysis. Methylation-specific PCR analysis of the P3H3 CpG island detects methylation in MDA MB 231, MDA MB 361, MDA MB 468, MCF7, BT474 and SKBR3, showing a clear correlation between methylation and down-regulation of mRNA. (D) Methylation-specific PCR analysis of P3H2 and P3H3 genes in ovarian carcinoma cell lines. The P3H3 CpG island is clearly methylated in OVCAR3, JAMA2 and IGROV cell lines. In contrast, there is no evidence of methylation in the P3H2 CpG island in any of the cell lines analysed.
Figure 2
Schematic representation of bisulphite sequence analysis of the P3H2 CpG island in breast carcinoma cell lines and normal breast epithelium (HMEC). Bisulphite sequencing was performed as described in Materials and methods. Primer pairs 1 and 2 used for MSP are indicated as set 1 and set 2 respectively. The thick black line on the scale indicates the position of exon 1 relative to the CpG island. The position of the part of the P3H2 open-reading frame within the CpG island is indicated by the broken line above the scale. Vertical lines below the scale represent individual CpG dinucleotides within the CpG island. The density of methylation for each cell line is represented by a quartile of blocks corresponding to each CpG. Black shading represents up to 25% methylation. Open blocks indicate no methylation. There is dense methylation in the MDA MB 361, MDA MB 453, MCF7, T47D and BT474 cell lines. There is no methylation in HMEC.
Figure 3
Schematic representation of bisulphite sequence analysis of the P3H3 CpG island in breast carcinoma cell lines and normal breast epithelium (HMEC). Bisulphite sequencing was performed as described in Materials and methods. Primers used for MSP are indicated above the scale. The thick black line on the scale indicates the position of exon 1 relative to the CpG island. The position of the part of the P3H3 open-reading frame within the CpG island is indicated by the broken line above the scale. Vertical lines below the scale represent individual CpG dinucleotides within the CpG island. The density of methylation for each cell line is represented by a quartile of blocks corresponding to each CpG. Black shading represents up to 25% methylation. Open blocks indicate no methylation. There is methylation in MDA MB 231, MDA MB 361, MDA MB 468, MCF7 and BT474. The CpG island is uniformly unmethylated in HMEC.
Figure 4
The P3H2 and P3H3 CpG islands are methylated in primary breast carcinomas. (A) Bisulphite sequence analysis of P3H2 CpG island in eight randomly selected primary breast carcinomas. The figure shows a schematic representation of the P3H2 CpG island as described in legend for Figure 2. Vertical lines below the scale represent individual CpG dinucleotides within the CpG island. The density of methylation for each cell line is represented by a quartile of blocks corresponding to each CpG. Black shading represents up to 25% methylation. Open blocks indicate no methylation. (B) Methylation-specific PCR analysis of P3H2 in primary breast carcinomas. It is shown with the number of each carcinoma analysed by both MSP and bisulphite sequencing indicated (underlined in the MSP gel). Unmethylated (CU) and methylated (CM) control DNAs, modified in parallel with primary cancer DNA samples in each case, are also shown. Cases 277, 424, 431, 446, 453 and 473 were identified as positive for methylation by MSP with primer set 2 as shown, whereas cases 301 and 326 were negative confirming the sensitivity and specificity of this primer set for methylation detection.
Figure 5
Ectopic expression of P3H2 and P3H3 in cells lacking endogenous expression suppresses cell proliferation. Expression plasmids for each gene or empty vector alone were introduced into individual cell lines as shown and transfected cells selected in G418. Surviving colonies were stained and counted after 16 days. (A) Expression of transfected plasmids for P3H2 and P3H3. T47D cells (which lack endogenous P3H2) and MDA MB 231 cells (which lack endogenous P3H3) were transfected with either control vector (C) or P3H2 and P3H3 expression plasmids respectively as indicated. Cell lysates were prepared and subjected to western blot analysis as described in Materials and methods. (B) Expression of transfected plasmids for P3H2 and P3H3 in MCF7 cells. MCF7 cells, which lack endogenous expression of both P3H2 and P3H3, were transfected with the indicated expression plasmids. Expression of transfected plasmids was analysed by RT–PCR as described in Materials and Methods. (C) Representative experiment showing suppression of colony growth by ectopic expression of P3H2 and P3H3. MCF7 cells (which lack endogenous expression of both P3H2 and P3H3) were transfected with expression plasmids for P3H2 and P3H3 as indicated. Cells were grown in the presence of G418 for 16 days, and surviving colonies were stained and counted. (D) Suppression of colony growth of MCF7, T47D and MDA MB 231 cells by ectopic expression of P3H2 and P3H3. The data shown are mean number of colonies (±standard error of mean) from three independent plates.
Similar articles
- Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers.
Yuan J, Luo RZ, Fujii S, Wang L, Hu W, Andreeff M, Pan Y, Kadota M, Oshimura M, Sahin AA, Issa JP, Bast RC Jr, Yu Y. Yuan J, et al. Cancer Res. 2003 Jul 15;63(14):4174-80. Cancer Res. 2003. PMID: 12874023 - Post-translationally abnormal collagens of prolyl 3-hydroxylase-2 null mice offer a pathobiological mechanism for the high myopia linked to human LEPREL1 mutations.
Hudson DM, Joeng KS, Werther R, Rajagopal A, Weis M, Lee BH, Eyre DR. Hudson DM, et al. J Biol Chem. 2015 Mar 27;290(13):8613-22. doi: 10.1074/jbc.M114.634915. Epub 2015 Feb 2. J Biol Chem. 2015. PMID: 25645914 Free PMC article. - Collagen prolyl hydroxylase 3 has a tumor suppressive activity in human lung cancer.
Li Y, Chen Y, Ma Y, Nenkov M, Haase D, Petersen I. Li Y, et al. Exp Cell Res. 2018 Feb 1;363(1):121-128. doi: 10.1016/j.yexcr.2017.12.020. Epub 2017 Dec 23. Exp Cell Res. 2018. PMID: 29277505 - Collagen Prolyl Hydroxylases Are Bifunctional Growth Regulators in Melanoma.
Atkinson A, Renziehausen A, Wang H, Lo Nigro C, Lattanzio L, Merlano M, Rao B, Weir L, Evans A, Matin R, Harwood C, Szlosarek P, Pickering JG, Fleming C, Sim VR, Li S, Vasta JT, Raines RT, Boniol M, Thompson A, Proby C, Crook T, Syed N. Atkinson A, et al. J Invest Dermatol. 2019 May;139(5):1118-1126. doi: 10.1016/j.jid.2018.10.038. Epub 2018 Nov 16. J Invest Dermatol. 2019. PMID: 30452903 Free PMC article. Review. - Epigenetic silencing of tumor suppressor genes in pancreatic cancer.
Lomberk GA. Lomberk GA. J Gastrointest Cancer. 2011 Jun;42(2):93-9. doi: 10.1007/s12029-011-9256-2. J Gastrointest Cancer. 2011. PMID: 21318291 Review.
Cited by
- Role of ZNF334 in cervical cancer: implications for EMT reversal and tumor suppression.
Li Q, Zhou X, Xiao J, Gong Y, Gong X, Shao B, Wang J, Zhao L, Xiong Q, Wu Y, Tang J, Yang Q, Tang J, Xiang T. Li Q, et al. Med Oncol. 2024 Jul 2;41(8):191. doi: 10.1007/s12032-024-02433-2. Med Oncol. 2024. PMID: 38954116 - Site-specific CpG methylation in the CCAAT/enhancer binding protein delta (CEBPδ) CpG island in breast cancer is associated with metastatic relapse.
Palmieri C, Monteverde M, Lattanzio L, Gojis O, Rudraraju B, Fortunato M, Syed N, Thompson A, Garrone O, Merlano M, Lo Nigro C, Crook T. Palmieri C, et al. Br J Cancer. 2012 Aug 7;107(4):732-8. doi: 10.1038/bjc.2012.308. Epub 2012 Jul 10. Br J Cancer. 2012. PMID: 22782348 Free PMC article. - Transcriptome-wide 1-methyladenosine functional profiling of messenger RNA and long non-coding RNA in bladder cancer.
Yin JJ, Song YL, Guo YF, Dai YH, Chang Q, Wang T, Sun GQ, Lu P, Song DK, Zhang LR. Yin JJ, et al. Front Genet. 2024 Feb 28;15:1333931. doi: 10.3389/fgene.2024.1333931. eCollection 2024. Front Genet. 2024. PMID: 38482382 Free PMC article. - Analysis of Tryptophan and Its Main Metabolite Kynurenine and the Risk of Multiple Cancers Based on the Bidirectional Mendelian Randomization Analysis.
Li R, Wang X, Zhang Y, Xu X, Wang L, Wei C, Liu L, Wang Z, Li Y. Li R, et al. Front Oncol. 2022 Apr 14;12:852718. doi: 10.3389/fonc.2022.852718. eCollection 2022. Front Oncol. 2022. PMID: 35494045 Free PMC article. - A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens.
Fernandes RJ, Farnand AW, Traeger GR, Weis MA, Eyre DR. Fernandes RJ, et al. J Biol Chem. 2011 Sep 2;286(35):30662-30669. doi: 10.1074/jbc.M111.267906. Epub 2011 Jul 11. J Biol Chem. 2011. PMID: 21757687 Free PMC article.
References
- Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, Pepin MG, Weis M, Eyre DR, Walsh J, Lambert D, Green A, Robinson H, Michelson M, Houge G, Lindman C, Martin J, Ward J, Lemyre E, Mitchell JJ, Krakow D, Rimoin DL, Cohn DH, Byers PH, Lee B (2008) CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat 29: 1435–1442 - PMC - PubMed
- Baylin SB (2005) DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2Suppl (1): S4–S11 - PubMed
- Baylin SB, Ohm JE (2006) Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107–116 - PubMed
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 - PubMed
- Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, Bulas DI, Kozma C, Smith PA, Eyre DR, Marini JC (2007) Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet 39: 359–365 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases