De novo generation of infectious prions in vitro produces a new disease phenotype - PubMed (original) (raw)
De novo generation of infectious prions in vitro produces a new disease phenotype
Marcelo A Barria et al. PLoS Pathog. 2009 May.
Erratum in
- PLoS Pathog. 2013 Mar;9(3). doi:10.1371/annotation/4b55946a-edb5-4feb-aeb5-2ee160394d17
Abstract
Prions are the proteinaceous infectious agents responsible for Transmissible Spongiform Encephalopathies. Compelling evidence supports the hypothesis that prions are composed exclusively of a misfolded version of the prion protein (PrP(Sc)) that replicates in the body in the absence of nucleic acids by inducing the misfolding of the cellular prion protein (PrP(C)). The most common form of human prion disease is sporadic, which appears to have its origin in a low frequency event of spontaneous misfolding to generate the first PrP(Sc) particle that then propagates as in the infectious form of the disease. The main goal of this study was to mimic an early event in the etiology of sporadic disease by attempting de novo generation of infectious PrP(Sc)in vitro. For this purpose we analyzed in detail the possibility of spontaneous generation of PrP(Sc) by the protein misfolding cyclic amplification (PMCA) procedure. Under standard PMCA conditions, and taking precautions to avoid cross-contamination, de novo generation of PrP(Sc) was never observed, supporting the use of the technology for diagnostic applications. However, we report that PMCA can be modified to generate PrP(Sc) in the absence of pre-existing PrP(Sc) in different animal species at a low and variable rate. De novo generated PrP(Sc) was infectious when inoculated into wild type hamsters, producing a new disease phenotype with unique clinical, neuropathological and biochemical features. Our results represent additional evidence in support of the prion hypothesis and provide a simple model to study the mechanism of sporadic prion disease. The findings also suggest that prion diversity is not restricted to those currently known, and that likely new forms of infectious protein foldings may be produced, resulting in novel disease phenotypes.
Conflict of interest statement
Dr. Soto is an inventor on the PMCA technology and a Founder, Vice-President and Chief Scientific Officer of Amprion Inc, a biotech company focusing on the development of early and sensitive diagnosis for prion diseases and other disorders involving protein misfolding.
Figures
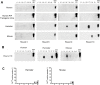
Figure 1. Standard PMCA does not lead to spontaneous generation of PrPSc.
Ten different mouse (A) and hamster (B) brain samples were subjected to serial rounds of PMCA in the absence of PrPSc inoculum. A total of 20 rounds of 144 PMCA cycles were done and after each round the appearance of PrPres after PK digestion was tested by western blot. The figure shows the results obtained in passages 6, 10, 16 and 20, and 5 representative samples of the 10 analyzed. In half of the samples, the PMCA reaction was done in the absence and the other half in the presence of 20 µg/ml synthetic poly-A oligonucleotide. These studies were done in a prion-free room using all new equipment and reagents. In some lanes (labeled with an asterisk) it is possible to appreciate a very faint band with the same molecular weight as PrPC, which is the result of incomplete digestion with PK. The efficiency of PMCA using these conditions was tested by running standard PMCA experiments using different dilutions of infected mouse (C) or hamster (D) brain homogenate. The experiment was done using 144 PMCA cycles and the results showed a robust amplification of the signal as previously described ,. All samples were digested with PK before western blot, except in the normal brain homogenate (NBH), used as control of PrPC migration. MBH: healthy mouse brain homogenate; HBH: healthy hamster brain homogenate; F: frozen samples; A: amplified samples.
Figure 2. De novo generation of PrPSc in diverse animal species.
(A) Samples from 10 different healthy brain homogenates from human, hamster, mouse or humanized transgenic mice were subjected to successive rounds of extended PMCA. Each round consisted of 240 PMCA cycles (30 min incubation followed by a 20 s pulse of sonication). Aliquots of each sample were treated with PK and analyzed by western blot. The results of rounds 3, 6, 9 and 10 are shown. (B) The same experiment as in A was repeated in a prion-free laboratory with all new equipment and reagents to reduce the possibility of cross-contamination. Only the round 10 is shown, but overall the results are very similar as those described in panel A. All samples were digested with PK before Western blot, except in the normal brain homogenate (NBH), used as control of PrPC migration. (C) Rate of spontaneous generation of protease-resistant PrP (PrPres) in different species at each of the PMCA rounds performed. Full bars represent the results obtained in the experiment conducted in our standard facility (as shown in panel A) and empty bars the experiment done in prion-free room with all new equipment and reagents (as shown in panel B).
Figure 3. De novo generated PrPSc is infectious, producing a new clinical disease.
(A) Groups of 5 hamsters were intra-cerebrally (i.c.) inoculated with similar quantities of PrPSc from either de novo generated protein (PGP-h1) or three well-established hamster prion strains, including 263K, HY and DY. Clinical signs were monitored weekly as described in Materials and Methods and when the signs progressed up to the level 4 of our scale, animals were considered sick and sacrificed to avoid excessive pain. This time is recorded as incubation period and is represented in the figure as days post-inoculation (d.p.i.). Differences on the incubation periods produced by the diverse sources of infectious material were analyzed by one-way ANOVA and were found highly significant (P<0.001). The significance of the differences between PGP-h1 and each of the other hamster prion strains were evaluated by the Dunnett Multiple Comparison post test. All differences were statistically significant (P<0.001). Behavioral alterations during the clinical phase of the disease in animals inoculated with various prion strains were assessed by an open field test, as described in Materials and Methods. Hamsters at mid stages of the disease (around 1 week after the first clinical signs were observed) were placed in a corner of the field and its behavior recorded for five minutes. We measured and analyzed total distance traveled during 20 s intervals (B), time that animals spent in vertical activity (C) and inactive time (D). The last two parameters are represented as a percentage of the total time. Tests were done twice for each animal and the result showed in the figure correspond to the average of the two determinations. Behavioral data was analyzed by one-way ANOVA with the Dunnett multiple comparison post-test. Statistical probability that the differences between PGP-h1 and control animals or hamsters inoculated with the other strains are different is indicated in the figure.
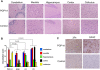
Figure 4. Histopathological brain damage in hamsters inoculated with de novo generated prions.
(A) Five different brain areas of animals sacrificed with clinical signs of disease induced by PGP-h1 infection and un-inoculated age-matched controls were analyzed histologically for spongiform degeneration after hematoxilin-eosin staining. (B) The vacuolation profile in each brain area was estimated using a semi-quantitative scale, as described in Materials and Methods. We also included in the analysis brain sections from animals inoculated with the other hamster prion strains studied. The values represent the average±standard error of the extent of vacuolation from the 5 animals analyzed in each set. Statistical analysis by two-ways ANOVA, using brain regions and prion origin as the variables indicated that differences were highly significant (P<0.001). To assess the significance of the differences between each known prion strain and PGP-h1, we used the Dunnett multiple comparison post-test and the P values for each combination are shown in the figure. (C) PrPSc accumulation and astroglyosis were studied by immunohistochemistry using anti-PrP and anti-GFAP antibodies, respectively. In the figure we show the staining in medulla as a representative brain region where substantial damage was observed. Staining of the brain of un-inoculated animals was included as a negative control.
Figure 5. Biochemical characteristics of PrPSc obtained in animals infected with de novo generated prions.
(A) To estimate the relative quantity of PrPSc in the brain of animals inoculated with different prion strains, the tissue was homogenized and aliquots corresponding to a 10-, 50-, and 1000-fold dilutions (respect to the entire brain) were analyzed by western blot after PK treatment. (B) The size of the PK-resistant fragment in each strain was assessed by western blot after treatment with the protease and deglycosylated as described in Materials and Methods. (C) The relative sensitivity of PGP-h1 PrPSc to proteolytic degradation was studied and compared with that of PrPSc associated to other hamster strains. For this study, aliquots of brain homogenate were incubated for 60 min at 37°C with the indicated concentrations of PK and PrPSc signal remaining was detected by Western blot. All samples were digested with PK before Western blot, except in the normal brain homogenate (NBH), used as control of PrPC migration. (D) Densitometric analysis of the western blots of 3 independent experiments as the one shown in the panel C was done to calculate the quantity of PrPSc digested with each PK concentration. This data enable to determine the susceptibility of PrPSc from the various sources to PK digestion and to estimate the PK50 value, which corresponds to the PK concentration needed to degrade 50% of the protein. The PK50 values (expressed in ug/ml of the protease) for each strain are indicated at the bottom of panel C. The data represent the average±standard error. The data was analyzed by two ways ANOVA (with source of the materials and PK concentration as the variables) and the Dunnett multiple comparison post-test. Each set of data was compared to the results obtained with the PGP-h1 strain and significant differences are highlighted with asterisks (* = P<0.05; ** = P<0.01; *** = P<0.001).
Similar articles
- Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions.
Castilla J, Gonzalez-Romero D, Saá P, Morales R, De Castro J, Soto C. Castilla J, et al. Cell. 2008 Sep 5;134(5):757-68. doi: 10.1016/j.cell.2008.07.030. Cell. 2008. PMID: 18775309 Free PMC article. - Sc237 hamster PrPSc and Sc237-derived mouse PrPSc generated by interspecies in vitro amplification exhibit distinct pathological and biochemical properties in tga20 transgenic mice.
Yoshioka M, Imamura M, Okada H, Shimozaki N, Murayama Y, Yokoyama T, Mohri S. Yoshioka M, et al. Microbiol Immunol. 2011 May;55(5):331-40. doi: 10.1111/j.1348-0421.2011.00328.x. Microbiol Immunol. 2011. PMID: 21362027 - De Novo Generation of a Unique Cervid Prion Strain Using Protein Misfolding Cyclic Amplification.
Meyerett-Reid C, Wyckoff AC, Spraker T, Pulford B, Bender H, Zabel MD. Meyerett-Reid C, et al. mSphere. 2017 Jan 25;2(1):e00372-16. doi: 10.1128/mSphere.00372-16. eCollection 2017 Jan-Feb. mSphere. 2017. PMID: 28144628 Free PMC article. - Protein misfolding cyclic amplification (PMCA): Current status and future directions.
Saá P, Cervenakova L. Saá P, et al. Virus Res. 2015 Sep 2;207:47-61. doi: 10.1016/j.virusres.2014.11.007. Epub 2014 Nov 13. Virus Res. 2015. PMID: 25445341 Review. - Protein Misfolding Cyclic Amplification of Infectious Prions.
Moda F. Moda F. Prog Mol Biol Transl Sci. 2017;150:361-374. doi: 10.1016/bs.pmbts.2017.06.016. Epub 2017 Jul 31. Prog Mol Biol Transl Sci. 2017. PMID: 28838669 Review.
Cited by
- Ethics in prion disease.
Bechtel K, Geschwind MD. Bechtel K, et al. Prog Neurobiol. 2013 Nov;110:29-44. doi: 10.1016/j.pneurobio.2013.07.001. Epub 2013 Jul 29. Prog Neurobiol. 2013. PMID: 23906487 Free PMC article. - Generation of prions in vitro and the protein-only hypothesis.
Diaz-Espinoza R, Soto C. Diaz-Espinoza R, et al. Prion. 2010 Apr-Jun;4(2):53-9. doi: 10.4161/pri.4.2.11960. Epub 2010 Apr 5. Prion. 2010. PMID: 20448454 Free PMC article. - Prion protein misfolding affects calcium homeostasis and sensitizes cells to endoplasmic reticulum stress.
Torres M, Castillo K, Armisén R, Stutzin A, Soto C, Hetz C. Torres M, et al. PLoS One. 2010 Dec 29;5(12):e15658. doi: 10.1371/journal.pone.0015658. PLoS One. 2010. PMID: 21209925 Free PMC article. - Prion seeded conversion and amplification assays.
Orrú CD, Caughey B. Orrú CD, et al. Top Curr Chem. 2011;305:121-33. doi: 10.1007/128_2011_184. Top Curr Chem. 2011. PMID: 21678135 Free PMC article. Review. - Factors affecting the accuracy of urine-based biomarkers of BSE.
Plews M, Lamoureux L, Simon SL, Graham C, Ruddat V, Czub S, Knox JD. Plews M, et al. Proteome Sci. 2011 Feb 7;9(1):6. doi: 10.1186/1477-5956-9-6. Proteome Sci. 2011. PMID: 21299878 Free PMC article.
References
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. - PubMed
- Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. - PubMed
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, et al. Synthetic mammalian prions. Science. 2004;305:673–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI77774/AI/NIAID NIH HHS/United States
- R01 NS049173/NS/NINDS NIH HHS/United States
- R01 NS49173/NS/NINDS NIH HHS/United States
- P01 AI077774/AI/NIAID NIH HHS/United States
- P01 AG14359/AG/NIA NIH HHS/United States
- P01 AG014359/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials