High-throughput bisulfite sequencing in mammalian genomes - PubMed (original) (raw)
High-throughput bisulfite sequencing in mammalian genomes
Zachary D Smith et al. Methods. 2009 Jul.
Abstract
DNA methylation is a critical epigenetic mark that is essential for mammalian development and aberrant in many diseases including cancer. Over the past decade multiple methods have been developed and applied to characterize its genome-wide distribution. Of these, reduced representation bisulfite sequencing (RRBS) generates nucleotide resolution DNA methylation bisulfite sequencing libraries that enrich for CpG-dense regions by methylation-insensitive restriction digestion. Here we provide an extensive, optimized protocol for generating RRBS libraries and discuss the power of this strategy for methylome profiling. We include information on sequence analysis and the relative coverage over genomic regions of interest for a representative mouse MspI generated RRBS library. Contemporary sequencing and array-based technologies are compared against sample throughput and coverage, highlighting the variety of options available to investigate methylation on the genome-scale.
Figures
Figure 1. Reduced Representation Bisulfite Sequencing
- General outline of RRBS library construction protocol including all key steps
- Overview of RRBS read alignment
- Sequencing coverage at selected regions of interest within the mouse genome as generated from a representative V6.5 mouse Embryonic Stem (ES) cell library.
*CpG Islands are as described in Ref. and extend beyond 700 bp in length.
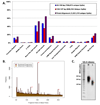
Figure 2. _Msp_I Generated RRBS Libraries from Mouse Genomic DNA
- Overlap of sequenced/aligned CpGs with specific regions of interest in the mouse genome for typical low (40–120bp) and high (120–220bp) fragment libraries (single CpGs can simultaneous overlap with more than one region class)
- Size distribution of in silico digested fragments compared to experimentally generated RRBS libraries (low and high are combined). Only unique sequences are aligned. Asterisks (*) denote redundant microsatellite fragments.
- Diagnostic gel of high (H) and low (L) fragment libraries generated from V6.5 mouse Embryonic Stem (ES) cells. Asterisks (*) correspond to microsatellite markers as predicted in silico.
Similar articles
- Double restriction-enzyme digestion improves the coverage and accuracy of genome-wide CpG methylation profiling by reduced representation bisulfite sequencing.
Wang J, Xia Y, Li L, Gong D, Yao Y, Luo H, Lu H, Yi N, Wu H, Zhang X, Tao Q, Gao F. Wang J, et al. BMC Genomics. 2013 Jan 16;14:11. doi: 10.1186/1471-2164-14-11. BMC Genomics. 2013. PMID: 23324053 Free PMC article. - Plant-RRBS, a bisulfite and next-generation sequencing-based methylome profiling method enriching for coverage of cytosine positions.
Schmidt M, Van Bel M, Woloszynska M, Slabbinck B, Martens C, De Block M, Coppens F, Van Lijsebettens M. Schmidt M, et al. BMC Plant Biol. 2017 Jul 6;17(1):115. doi: 10.1186/s12870-017-1070-y. BMC Plant Biol. 2017. PMID: 28683715 Free PMC article. - Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling.
Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Gu H, et al. Nat Protoc. 2011 Apr;6(4):468-81. doi: 10.1038/nprot.2010.190. Epub 2011 Mar 18. Nat Protoc. 2011. PMID: 21412275 - Methods for CpG Methylation Array Profiling Via Bisulfite Conversion.
Leti F, Llaci L, Malenica I, DiStefano JK. Leti F, et al. Methods Mol Biol. 2018;1706:233-254. doi: 10.1007/978-1-4939-7471-9_13. Methods Mol Biol. 2018. PMID: 29423802 Free PMC article. Review. - Methodological aspects of whole-genome bisulfite sequencing analysis.
Adusumalli S, Mohd Omar MF, Soong R, Benoukraf T. Adusumalli S, et al. Brief Bioinform. 2015 May;16(3):369-79. doi: 10.1093/bib/bbu016. Epub 2014 May 27. Brief Bioinform. 2015. PMID: 24867940 Review.
Cited by
- Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing.
Guo H, Zhu P, Guo F, Li X, Wu X, Fan X, Wen L, Tang F. Guo H, et al. Nat Protoc. 2015 May;10(5):645-59. doi: 10.1038/nprot.2015.039. Epub 2015 Apr 2. Nat Protoc. 2015. PMID: 25837417 - Current and Emerging Technologies for the Analysis of the Genome-Wide and Locus-Specific DNA Methylation Patterns.
Tost J. Tost J. Adv Exp Med Biol. 2022;1389:395-469. doi: 10.1007/978-3-031-11454-0_16. Adv Exp Med Biol. 2022. PMID: 36350519 - In silico analysis identifies novel restriction enzyme combinations that expand reduced representation bisulfite sequencing CpG coverage.
Martinez-Arguelles DB, Lee S, Papadopoulos V. Martinez-Arguelles DB, et al. BMC Res Notes. 2014 Aug 15;7:534. doi: 10.1186/1756-0500-7-534. BMC Res Notes. 2014. PMID: 25127888 Free PMC article. - Universal endogenous gene controls for bisulphite conversion in analysis of plant DNA methylation.
Wang J, Wang C, Long Y, Hopkins C, Kurup S, Liu K, King GJ, Meng J. Wang J, et al. Plant Methods. 2011 Dec 2;7:39. doi: 10.1186/1746-4811-7-39. Plant Methods. 2011. PMID: 22132777 Free PMC article. - Genomic distribution and inter-sample variation of non-CpG methylation across human cell types.
Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, Gnirke A, Meissner A. Ziller MJ, et al. PLoS Genet. 2011 Dec;7(12):e1002389. doi: 10.1371/journal.pgen.1002389. Epub 2011 Dec 8. PLoS Genet. 2011. PMID: 22174693 Free PMC article.
References
- Bestor TH. Hum. Mol. Genet. 2000;9:2395–2402. - PubMed
- Bird A. Genes Dev. 2002;16:6–21. - PubMed
- Bernstein BE, Meissner A, Lander ES. Cell. 2007;128:669–681. - PubMed
- Santos F, Hendrich B, Reik W, et al. Dev. Biol. 2002;241:172–182. - PubMed
- Reik W, Dean W, Walter J. Science. 2001;293:1089–1093. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous