Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity - PubMed (original) (raw)
Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity
Ramesh Rathinakumar et al. J Am Chem Soc. 2009.
Abstract
We recently described 10 peptides selected from a 16,384-member combinatorial library based on their ability to permeabilize synthetic lipid vesicles in vitro. These peptides did not share a common sequence motif, length, or net charge; nonetheless, they shared a mechanism of action that is similar to the natural membrane permeabilizing antimicrobial peptides (AMP). To characterize the selected peptides and to compare the activity of AMPs in vivo and in vitro, we report on the biological activity of the same selected peptides in bacteria, fungi, and mammalian cells. Each of the peptides has sterilizing activity against all classes of microbes tested, at 2-8 microM peptide, with only slight hemolytic or cytotoxicity against mammalian cells. Similar to many natural AMPs, bacteria are killed within a few minutes of peptide addition, and the lethal step in vivo is membrane permeabilization. Single D-amino acid substitutions eliminated or diminished the secondary structure of the peptides, and yet, they retained activity against some microbes. Thus, secondary structure and biological activity are not coupled, consistent with the hypothesis that AMPs do not form pores of well-defined structure in membranes but rather destabilize membranes by partitioning into membrane interfaces and disturbing the organization of the lipids, a property that we have called "interfacial activity". The observation that broad-spectrum activity, but not all antimicrobial activity, is lost by small changes to the peptides suggests that the in vitro screen is specifically selecting for the rare peptides that have broad-spectrum activity. We put forth the hypothesis that methods focusing on screening peptide libraries in vitro for members with the appropriate interfacial activity can enable the design, selection, and discovery of novel, potent, and broad-spectrum membrane-active antibiotics.
Figures
Figure 1
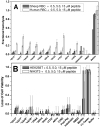
Figure 1A. Membrane-active peptides from library Figure 1B. D-amino acid substituted membrane-active peptides A: The peptides used in this study. These peptides were selected from a 16,384 member combinatorial peptide library using an in vitro high-throughput screen for the ability to permeabilize synthetic lipid vesicles containing 90% phosphatidylcholine lipids and 10% phosphatidylglycerol lipids. Library design, peptide selection and characterization of the active peptides in vitro are described elsewhere,,. The combinatorial sites in the peptides are highlighted in red. The RRG- and –GRR terminal cassettes were also varied combinatorially. On the right side, we give the four letter codes we use herein to represent the peptides. The code is based on the identity of the amino acids in the four combinatorial sites. The asterisk symbols (*) represents the terminal cassettes, when present. B: The D-amino acid substituted peptide homologs used in this study. Four of the peptides above were synthesized with a single D-leucine substitution near the C-terminus to study the coupling of structure to biological activity. The D-leucine is represented by ‘dL’ in blue.
Figure 2. Antimicrobial activity
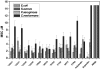
The sterilizing antimicrobial activity of the ten peptides against Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Cryptococcus neoformans. Each row in a 96-well plate contained one peptide in serial dilution from 10 μM to 0.1 μM. Cells at 103 cells/ml suspended in minimal media were added and incubated for 3 hrs, and then 2× growth media is added to the cells which were allowed to recover overnight at 37 °C. Wells containing less than a threshold peptide concentration were opaque, indicating stationary phase growth, while wells containing above the threshold peptide concentration were transparent, indicating no growth at all. The lowest concentration of peptide that prevented growth is the minimal sterilizing concentration (MSC). MSC values in the figure are the average of 3-5 independent measurements with standard errors.
Figure 3. Hemolytic and cytotoxic activity
Effect of selected peptides on mammalian cell membranes. A: Hemolytic activity of the selected peptides against sheep and human erythrocytes. Sheep erythrocytes (7.7 × 106 cells/ml) and human erythrocytes (2 × 107 cells/ml) in phosphate-sodium buffer solution (PBS) pH 7.4, were incubated with peptides at 0.5, 5 and 15 μM for 1 hour. For each peptide six values are shown, 0.5, 5 and 15 μM peptide for sheep and human erythrocytes, left to right. Cells were then centrifuged at 4000g for 2 minutes. The optical absorbance of hemoglobin at 540nm is used to measure hemolysis. Activity is expressed as percent hemolysis where zero is a buffer only control and 100% hemolysis is the value for osmotic lysis with distilled water. B: The cytotoxicity activity of the selected peptides against mammalian HEK293T and NIH3T3 cells. The cells at 5 × 105 cells/mL were treated with peptide at 0.5, 5 and 15 μM and incubated for 72 hours. Cell viability was measured by using the MTT assay in which mitochondrial reductases in living cells produce formazan crystals after the addition of MTT. The crystals were dissolved in isopropanol and the optical density measured at 550 nm. Activity is expressed as percent cytotoxicity where zero is a buffer-only control and 100% is the value for treatment with 1% of the detergent TWEEN.
Figure 4. Mitochondrial membrane permeabilization
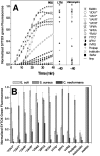
Microbial membrane permeabilization caused by the selected peptides. A: Cytoplasmic membrane permeabilization of E. coli measured by entry of the DNA binding dye SYTOX Green. Dye (1 μM) was added to 2 × 107 cells/ml suspended in PBS and the time course of fluorescence was monitored after addition of 5 μM peptide. Fluorescence, monitored with excitation at 485nm and emission at 520 nm, increases only when the dye can cross the membrane and bind DNA. The pore-forming peptide melittin served as a positive control for membrane permeabilization. The dye uptake is measured at three different assay conditions of E. coli. PBS: metabolically dormant cells in PBS buffer alone; LTM: metabolically active cells in “liquid test medium”, containing 1% growth medium in PBS; and valinomycin: cells treated with 40 μM valinomycin to dissipate the membrane potential. SYTOX Green fluorescence is shown relative to that caused by complete lysis with melittin at 40 minutes. B: Membrane permeabilization of different microorganisms caused by the selected peptides. Membrane permeabilization was measured using the SYTOX Green DNA binding dye as described above. Cells in PBS (2 × 107 cells/mL) were incubated with 1 μM SYTOX Green and 5 μM peptide. After 40 minutes, the net uptake of the dye through the plasma membrane was monitored by fluorescence and normalized to the fluorescence in the presence of the same amount of melittin.
Figure 5. Microbial membrane depolarization
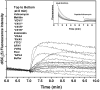
Cytoplasmic membrane depolarization of E. coli. A membrane potential-sensitive dye, 3,3′- dipropylthiacarbocyanine [diSC3(5)] was added to E. coli cells (2 × 107 cells/mL) at 50 nM and the change in dye fluorescence (excitation at 622 nm, emission at 670 nm) due to membrane binding and self quenching was monitored until it was stable (inset). Peptides (5 μM) were added at 7 min and the loss of membrane potential was measured. The fluorescence increase intensity for complete collapse of cytoplasm membrane potential is given by the valinomycin curve, which is closely matched by the positive control peptide melittin.
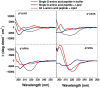
Figure 6. Circular dichroism spectroscopy of D-amino acid substituted peptides
Change in secondary structure content of D-leucine substituted peptides. Circular dichroism spectra of the single D-amino acid substituted peptides at 50 μM concentration in phosphate buffer, pH 7.0. Spectra were taken before and after the addition of 2.5 mM phospholipid vesicles composed of 90% POPC and 10% POPG. The CD spectra of the same all L-peptide in phosphate buffer with 2.5 mM phospholipid vesicles are also shown for comparison. The ellipticity values are given as molar, or mean residue, ellipticities.
Similar articles
- High-throughput discovery of broad-spectrum peptide antibiotics.
Rathinakumar R, Wimley WC. Rathinakumar R, et al. FASEB J. 2010 Sep;24(9):3232-8. doi: 10.1096/fj.10-157040. Epub 2010 Apr 21. FASEB J. 2010. PMID: 20410445 Free PMC article. - Antimicrobial properties of membrane-active dodecapeptides derived from MSI-78.
Monteiro C, Fernandes M, Pinheiro M, Maia S, Seabra CL, Ferreira-da-Silva F, Costa F, Reis S, Gomes P, Martins MC. Monteiro C, et al. Biochim Biophys Acta. 2015 May;1848(5):1139-46. doi: 10.1016/j.bbamem.2015.02.001. Epub 2015 Feb 10. Biochim Biophys Acta. 2015. PMID: 25680229 - In silico design of polycationic antimicrobial peptides active against Pseudomonas aeruginosa and Staphylococcus aureus.
Hincapié O, Giraldo P, Orduz S. Hincapié O, et al. Antonie Van Leeuwenhoek. 2018 Oct;111(10):1871-1882. doi: 10.1007/s10482-018-1080-2. Epub 2018 Apr 6. Antonie Van Leeuwenhoek. 2018. PMID: 29626331 - Alpha-helical cationic antimicrobial peptides: relationships of structure and function.
Huang Y, Huang J, Chen Y. Huang Y, et al. Protein Cell. 2010 Feb;1(2):143-52. doi: 10.1007/s13238-010-0004-3. Epub 2010 Feb 6. Protein Cell. 2010. PMID: 21203984 Free PMC article. Review. - Describing the mechanism of antimicrobial peptide action with the interfacial activity model.
Wimley WC. Wimley WC. ACS Chem Biol. 2010 Oct 15;5(10):905-17. doi: 10.1021/cb1001558. ACS Chem Biol. 2010. PMID: 20698568 Free PMC article. Review.
Cited by
- BING, a novel antimicrobial peptide isolated from Japanese medaka plasma, targets bacterial envelope stress response by suppressing cpxR expression.
Dong M, Kwok SH, Humble JL, Liang Y, Tang SW, Tang KH, Tse MK, Lei JH, Ramalingam R, Koohi-Moghadam M, Au DWT, Sun H, Lam YW. Dong M, et al. Sci Rep. 2021 Jun 9;11(1):12219. doi: 10.1038/s41598-021-91765-4. Sci Rep. 2021. PMID: 34108601 Free PMC article. - Proteomic Response of Bacillus subtilis Spores under High Pressure Combined with Moderate Temperature and Random Peptide Mixture LK Treatment.
Pang Y, Wu R, Cui T, Zhang Z, Dong L, Chen F, Hu X. Pang Y, et al. Foods. 2022 Apr 13;11(8):1123. doi: 10.3390/foods11081123. Foods. 2022. PMID: 35454710 Free PMC article. - Scorpion Venom Antimicrobial Peptides Induce Caspase-1 Dependant Pyroptotic Cell Death.
Elrayess RA, Mohallal ME, Mobarak YM, Ebaid HM, Haywood-Small S, Miller K, Strong PN, Abdel-Rahman MA. Elrayess RA, et al. Front Pharmacol. 2022 Jan 10;12:788874. doi: 10.3389/fphar.2021.788874. eCollection 2021. Front Pharmacol. 2022. PMID: 35082671 Free PMC article. - Random peptide mixtures as new crop protection agents.
Topman S, Tamir-Ariel D, Bochnic-Tamir H, Stern Bauer T, Shafir S, Burdman S, Hayouka Z. Topman S, et al. Microb Biotechnol. 2018 Nov;11(6):1027-1036. doi: 10.1111/1751-7915.13258. Epub 2018 Feb 28. Microb Biotechnol. 2018. PMID: 29488347 Free PMC article. - Broad-Spectrum Antiviral Entry Inhibition by Interfacially Active Peptides.
Hoffmann AR, Guha S, Wu E, Ghimire J, Wang Y, He J, Garry RF, Wimley WC. Hoffmann AR, et al. J Virol. 2020 Nov 9;94(23):e01682-20. doi: 10.1128/JVI.01682-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32907984 Free PMC article.
References
- D'Agata EM. Rapidly rising prevalence of nosocomial multidrag-resistant, Gram-negative bacilli: a 9-year surveillance study. Infect Control Hosp Epidemiol. 2004;25(10):842–846. - PubMed
- Hancock RE. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1(3):156–164. - PubMed
- Avrahami D, Shai Y. Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry. 2002;41(7):2254–2263. - PubMed
- Makovitzki A, Shai Y. pH-dependent antifungal lipopeptides and their plausible mode of action. Biochemistry. 2005;44(28):9775–9784. - PubMed
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature (London) 2002;415(6870):389–395. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources