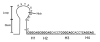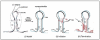Replication of porcine circoviruses - PubMed (original) (raw)
Review
Replication of porcine circoviruses
Florence Faurez et al. Virol J. 2009.
Abstract
Porcine circoviruses are circular single-stranded DNA viruses that infect swine and wild boars. Two species of porcine circoviruses exist. Porcine circovirus type 1 is non pathogenic contrary to porcine circovirus type 2 which is associated with the disease known as Post-weaning Multisystemic Wasting Syndrome. Porcine circovirus DNA has been shown to replicate by a rolling circle mechanism. Other studies have revealed similar mechanisms of rolling-circle replication in plasmids and single-stranded viruses such as Geminivirus. Three elements are important in rolling-circle replication: i) a gene encoding initiator protein, ii) a double strand origin, and iii) a single strand origin. However, differences exist between viruses and plasmids and between viruses. Porcine circovirus replication probably involves a "melting pot" rather than "cruciform" rolling-circle mechanism.This review provides a summary of current knowledge of replication in porcine circoviruses as models of the Circovirus genus. Based on various studies, the factors affecting replication are defined and the mechanisms involved in the different phases of replication are described or proposed.
Figures
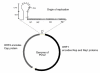
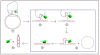
Figure 1
Representation of genome of PCV-2. Black arrow: Open Reading Frame 1 (ORF1), located on the positive strand, which encodes Rep and Rep' protein. Grey arrow: Open Reading Frame 2 (ORF2), located on the negative strand, which encodes Cap protein. Between ORF1 and ORF2 are intergenic regions. The origin of replication is located in the intergenic region between the beginnings of the two ORFs. H1, H2, H3 and H4 are hexamers.
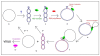
Figure 2
Rolling circle replication model of single strand viruses. Black strand: positive parental strand and viral genome. Blue strand: negative lagging strand. Red strand: positive leading strand. a and b: the conversion of the single strand viral genome in the double strand DNA replicative intermediate is produced in the cell. c and d: REP complex binds to the stem-loop structure which is the origin of replication and initiates the replication by nicking the DNA. d and e: cellular DNA polymerase initiates viral DNA replication from the free 3'OH extremity and the REP complex still binds the 5' extremity. f and g: after a round of replication, the REP complex closes the DNA and releases a single strand DNA and a double strand DNA. h', h" and h"': DNA can be used for replication or can be encapsidated.
Figure 3
PCV-2 replication origin. H1: Hexamer 1; H2: Hexamer 2; H3: Hexamer 3; H4: Hexamer 4 Blue nucleic acids: pentamers CACCT.
Figure 4
3D structure of the Rep protein N-term part of PCV-2. Left panel: Gibbon representation. In blue: 5 β sheets; red and green: α helix; grey: loop; yellow: α3 helix bearing the catalytic site. Right panel: zoom on β sheets and catalytic α helix. Blue amino acids: motif I (FTLN) of β2 sheet; Green amino acids: motif II (HLQGF) of β4 sheet; Red amino acids: motif III (YCSK). Reprinted from Journal of Molecular Biology, 367(2), Vega-Rocha S et al, "Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2.", pages 473–487, Copyright© 2007 [18], with permission from Elsevier.
Figure 5
Melting pot rolling circle replication model. i. Replication origin representation after binding of the Rep. Strands (+) and (-) are close together. The destabilized environment known as "melting pot" is included in the dotted oval. ii. Schematic representation of the template DNA strands during the initiation of DNA synthesis. The leading strand shifts strand 'a', and uses strands 'a' and 'b' as template DNA strands. iii. Schematic representation of the template DNA strands at the termination of DNA synthesis. The leading strand shifts strand 'b' and uses either the newly synthesized 'aN ' or strand 'b'. Black: positive polarized genome (+); Blue: negative strand; Red: potential base pairs representation. Reprinted from Journal of Virology 78(8), Cheung AK, "Detection of template strand switching during initiation and termination of DNA replication of porcine circovirus." pages 4268–4277, Copyright© 2004 [6], with permission from American Society for Microbiology.
Figure 6
Model of the termination of rolling circle replication described by Novick in the nineties. Black: parental strand; Red: newly synthesized strand; Blue: newly synthesized strand during a second round of replication. Step 1: After the first round of replication, the REP complex bound on the 5' end would be situated behind the generated stem-loop. Step 2: the tyrosine (Y) of the unused sub-unit during the initiation might cleave the regenerated replication origin. It will (then bind to the 5' end of the newly synthesized strand. Step 3: the 3'OH free end generated during step 2 and belonging to the parental strand might then exert a nucleophilic attack on the tyrosylphosphodiester bridge generated during the initiation step. This reaction would lead to the release of a single stranded DNA. Step 4: the tyrosine previously involved in initiation of the replication attacks the cutting site of the newly synthesized strand. This reaction would generate a free 3'OH on the newly synthesized strand and a tyrosylphosphodiester bridge with a 12 mers oligonucleotide. Steps 5 and 6: the free 3'OH generated at step 4 can attack the tyrosylphosphodiester bridge generated at step 2, that would close the double stranded DNA and release an inactivated Rep homodimer due to the binding of one tyrosine to one oligonucleotide. Adapted from Microbiology and Molecular Biology Reviews 62(2), del Solar G et al, "Replication and control of circular bacterial plasmids." pages 434–464, Copyright© 1998 [5], with permission from American Society for Microbiology.
Similar articles
- Identification of a protein essential for replication of porcine circovirus.
Mankertz A, Mankertz J, Wolf K, Buhk HJ. Mankertz A, et al. J Gen Virol. 1998 Feb;79 ( Pt 2):381-4. doi: 10.1099/0022-1317-79-2-381. J Gen Virol. 1998. PMID: 9472624 - Structural Perspectives of Beak and Feather Disease Virus and Porcine Circovirus Proteins.
Nath BK, Das S, Roby JA, Sarker S, Luque D, Raidal SR, Forwood JK. Nath BK, et al. Viral Immunol. 2021 Jan-Feb;34(1):49-59. doi: 10.1089/vim.2020.0097. Epub 2020 Dec 4. Viral Immunol. 2021. PMID: 33275868 Review. - Detection of template strand switching during initiation and termination of DNA replication of porcine circovirus.
Cheung AK. Cheung AK. J Virol. 2004 Apr;78(8):4268-77. doi: 10.1128/jvi.78.8.4268-4277.2004. J Virol. 2004. PMID: 15047840 Free PMC article. - Porcine circovirus: transcription and DNA replication.
Cheung AK. Cheung AK. Virus Res. 2012 Mar;164(1-2):46-53. doi: 10.1016/j.virusres.2011.10.012. Epub 2011 Oct 20. Virus Res. 2012. PMID: 22036834 Review.
Cited by
- Computational based design and tracking of synthetic variants of Porcine circovirus reveal relations between silent genomic information and viral fitness.
Baron L, Atar S, Zur H, Roopin M, Goz E, Tuller T. Baron L, et al. Sci Rep. 2021 May 19;11(1):10620. doi: 10.1038/s41598-021-89918-6. Sci Rep. 2021. PMID: 34012100 Free PMC article. - The development of a rapid SYBR Green I-based quantitative PCR for detection of Duck circovirus.
Wan C, Huang Y, Cheng L, Fu G, Shi SH, Chen H, Peng C, Lin F, Lin J. Wan C, et al. Virol J. 2011 Oct 7;8:465. doi: 10.1186/1743-422X-8-465. Virol J. 2011. PMID: 21978576 Free PMC article. - Canine circovirus: An emerging or an endemic undiagnosed enteritis virus?
Gomez-Betancur D, Vargas-Bermudez DS, Giraldo-Ramírez S, Jaime J, Ruiz-Saenz J. Gomez-Betancur D, et al. Front Vet Sci. 2023 Apr 17;10:1150636. doi: 10.3389/fvets.2023.1150636. eCollection 2023. Front Vet Sci. 2023. PMID: 37138920 Free PMC article. Review. - The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province.
Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, Peng Z, Xu S, Fu F, Yu J, Sun W, Xu S, Li J, Wang J. Zheng S, et al. Transbound Emerg Dis. 2017 Oct;64(5):1337-1341. doi: 10.1111/tbed.12667. Epub 2017 Jun 26. Transbound Emerg Dis. 2017. PMID: 28653486 Free PMC article. - Novel single-stranded, circular DNA virus identified in cats in Japan.
Takano T, Yanai Y, Hiramatsu K, Doki T, Hohdatsu T. Takano T, et al. Arch Virol. 2018 Dec;163(12):3389-3393. doi: 10.1007/s00705-018-4020-6. Epub 2018 Sep 14. Arch Virol. 2018. PMID: 30218220 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources