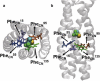Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, III: Molecular dynamics simulation incorporating a cyanophenylalanine spectroscopic probe - PubMed (original) (raw)
Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, III: Molecular dynamics simulation incorporating a cyanophenylalanine spectroscopic probe
Hongling Zou et al. Biophys J. 2009.
Abstract
A nitrile-derived amino acid, Phe(CN), has been used as an internal spectroscopic probe to study the binding of an inhalational anesthetic to a model membrane protein. The infrared spectra from experiment showed a blue-shift of the nitrile vibrational frequency in the presence of the anesthetic halothane. To interpret the infrared results and explore the nature of the interaction between halothane and the model protein, all-atom molecular dynamics (MD) simulations have been used to probe the structural and dynamic properties of the protein in the presence and absence of one halothane molecule. The frequency shift analyzed from MD simulations agrees well with the experimental infrared results. Decomposition of the forces acting on the nitrile probes demonstrates an indirect impact on the probes from halothane, namely a change of the protein's electrostatic local environment around the probes induced by halothane. Although the halothane remains localized within the designed hydrophobic binding cavity, it undergoes a significant amount of translational and rotational motion, modulated by the interaction of the trifluorine end of halothane with backbone hydrogens of the residues forming the cavity. This dominant interaction between halothane and backbone hydrogens outweighs the direct interaction between halothane and the nitrile groups, making it a good "spectator" probe of the halothane-protein interaction. These MD simulations provide insight into action of anesthetic molecules on the model membrane protein, and also support the further development of nitrile-labeled amino acids as spectroscopic probes within the designed binding cavity.
Figures
Figure 1
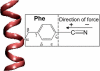
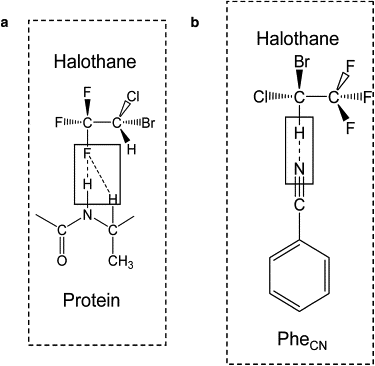
Illustration of the chemical structure of nonbiological amino acid PheCN and the definition of the nitrile bond axis. The helix to which the PheCN residue is attached is shown in ribbon representation.
Figure 2
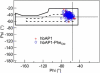
Ramachandran plot for residues 2–36 in all the helices of the apo protein hbAP1 (cross) and its mutant hbAP1-PheCN (circle) at the water-octane interface. The backbone conformation angles phi and psi are calculated as averages over the last 1 ns of the simulation. The dashed perimeter corresponds to the most favorable combinations of phi/psi values for a right-handed _α_-helical conformation, and the solid perimeter highlights the somewhat larger “Ramachandran-allowed” region.
Figure 3
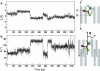
Time evolution of the halothane's translational (a) and rotational (b) motion within the hbAP1-PheCN bundle. The halothane's translational displacement Z is defined as the distance from the center of the halothane molecule (Z) to the center of the four Ala19 residues (O) projected along the peptide bundle long axis. The halothane's rotational angle θ is defined as the angle between the C-C axis of the halothane molecule and the peptide bundle long axis.
Figure 4
Instantaneous configuration of the hbAP1-PheCN with the halothane binding to the cavity viewed from the top (a) and from the side (b). The peptide is shown in ribbon representation and the four PheCN residues in wire-frame representation.
Figure 5
Two possibilities were considered for hydrogen-bond formation. (a) hydrogen-bond formation between the halothane trifluorine atoms and the backbone hydrogen atoms. (b) Hydrogen-bond formation between the acidic hydrogen of the halothane and the nitrile nitrogen of the PheCN residue.
Figure 6
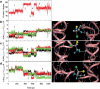
Correlations between the halothane's rotational motion (a) and the distance of approach of its relevant hydrogen-bond acceptor atoms to the cavity's backbone hydrogen atoms (b–d), which is shown in Fig. 5_a_. (a) Halothane's rotational angle θ is defined as in Fig. 3. (b–d) The distance between the halothane trifluorines (three fluorines are shown in three different colors) and the adjacent Ala (Ala139 in b and c, Ala99 in d) backbone amide-H (HN) or C_α_-H (H_α_). In a, the numbers 1–5 denotes the different phases of the halothane's orientation. Each phase is then matched well to the interaction between the halothane and the cavity backbone hydrogen atoms shown in the right panels of b–d. In the right panels, hydrogen bonds between the trifluorines and the HN or the H_α_ are represented in white dashed lines. Particularly, in right panel c, the same HN also forms a hydrogen bond with the backbone carbonyl oxygen, which is responsible for stabilizing the protein secondary structure. The peptide is shown in ribbon representation with backbone atoms in wire-frame representation. Nitrogen is colored in blue, oxygen in red, and carbon in silver. The halothane molecule is shown in CPK representation with trifluorines colored in cyan, bromine in yellow, chlorine in green, and hydrogen in white.
Figure 7
For two parallel simulations (simulation I, a and b; simulation II, c and d), correlations between the halothane's rotational motion (a and c) and the distance of approach of its relevant hydrogen-bond donor to the PheCN residues' nitrile-nitrogen (b and d), as presented in Fig. 5_b_, were studied with partial charges on nitrile groups included (left four panels) or removed (right four panels). (a and c) The halothane's orientational angle θ is defined as in Fig. 3. (b and d) The distance between acidic hydrogen of the halothane and the nitrile-nitrogen of the four PheCN residues are plotted, with each of them in a different gray scale.
Figure 8
Distribution of the aromatic ring's motion for two PheCN residues in the absence (gray bar) and presence of the halothane (black bar). χ is defined as the dihedral angle for C_α_-C_β_-C_γ_-C_δ_, characterizing the “flipping” motion of the aromatic ring; θ is defined as the angle between C_β_-C_γ_ and the bundle long axis, characterizing the “tilting” motion of the aromatic ring. The residues in consideration are two PheCN residues that are closer to the halothane. The addition of halothane seems to have no apparent effect on the widths of the distributions for either of these motions of aromatic rings for either of the two PheCN residues, indicating the absence of an interaction between halothane and aromatic rings of PheCN probes.
Figure 9
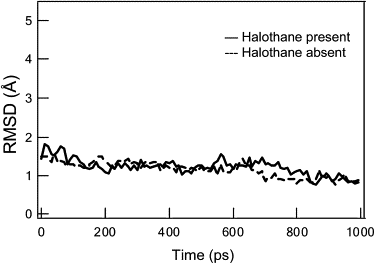
Halothane's impact on the backbone structure of the hbAP1-PheCN peptide bundle. Shown here is a comparison of the computed RMSD of backbone C_α_ atoms in the absence (dashed line) and presence (solid line) of the halothane molecule, respectively. The equation for calculating RMSD is as follows: RMSD=[1M∑i=1M(Ri−Rieq)2]1/2, where Ri and Rieq are the coordinates of the i_th backbone C_α of an instantaneous structure and the equilibrated structure, respectively.
Figure 10
Comparison of the backbone dynamics of the hbAP1-PheCN peptide bundle in the absence (dashed line) and presence (solid line) of halothane. Shown here is the computed RMSF of backbone C_α_ atoms for each residue. The four helices in the bundle are named H1, H2, H3, and H4. H1 and H2 are within one dimer; H3 and H4 belong to the other dimer. The equation for calculating RMSF is as follows: RMSF=[1T∑tj=1T(Ri(tj)−Ri¯)2]1/2, where Ri(tj) and Ri¯ are the coordinates of the i_th backbone C_α of an instantaneous structure at time tj and the coordinates averaged over time T, respectively. Note that the difference between RMSD and RMSF is that with the later the average is taken over time T, giving a value for each residue i; with RMSD the average is taken over the M residues, giving time specific values.
Similar articles
- Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, II: Fluorescence and vibrational spectroscopy using a cyanophenylalanine probe.
Liu J, Strzalka J, Tronin A, Johansson JS, Blasie JK. Liu J, et al. Biophys J. 2009 May 20;96(10):4176-87. doi: 10.1016/j.bpj.2009.01.055. Biophys J. 2009. PMID: 19450488 Free PMC article. - Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, I: Structural investigations via X-ray reflectivity from Langmuir monolayers.
Strzalka J, Liu J, Tronin A, Churbanova IY, Johansson JS, Blasie JK. Strzalka J, et al. Biophys J. 2009 May 20;96(10):4164-75. doi: 10.1016/j.bpj.2009.01.053. Biophys J. 2009. PMID: 19450487 Free PMC article. - Large-scale molecular dynamics simulations of general anesthetic effects on the ion channel in the fully hydrated membrane: the implication of molecular mechanisms of general anesthesia.
Tang P, Xu Y. Tang P, et al. Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16035-40. doi: 10.1073/pnas.252522299. Epub 2002 Nov 18. Proc Natl Acad Sci U S A. 2002. PMID: 12438684 Free PMC article. - Computational studies on the interactions of inhalational anesthetics with proteins.
Vemparala S, Domene C, Klein ML. Vemparala S, et al. Acc Chem Res. 2010 Jan 19;43(1):103-10. doi: 10.1021/ar900149j. Acc Chem Res. 2010. PMID: 19788306 Review. - Nitrile groups as vibrational probes of biomolecular structure and dynamics: an overview.
Lindquist BA, Furse KE, Corcelli SA. Lindquist BA, et al. Phys Chem Chem Phys. 2009 Oct 1;11(37):8119-32. doi: 10.1039/b908588b. Epub 2009 Jul 31. Phys Chem Chem Phys. 2009. PMID: 19756266 Review.
Cited by
- 5-Cyanotryptophan as an Infrared Probe of Local Hydration Status of Proteins.
Waegele MM, Tucker MJ, Gai F. Waegele MM, et al. Chem Phys Lett. 2009 Sep 1;478(4):249-253. doi: 10.1016/j.cplett.2009.07.058. Chem Phys Lett. 2009. PMID: 20161057 Free PMC article. - Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, II: Fluorescence and vibrational spectroscopy using a cyanophenylalanine probe.
Liu J, Strzalka J, Tronin A, Johansson JS, Blasie JK. Liu J, et al. Biophys J. 2009 May 20;96(10):4176-87. doi: 10.1016/j.bpj.2009.01.055. Biophys J. 2009. PMID: 19450488 Free PMC article. - New insights into the molecular mechanisms of general anaesthetics.
Chau PL. Chau PL. Br J Pharmacol. 2010 Sep;161(2):288-307. doi: 10.1111/j.1476-5381.2010.00891.x. Br J Pharmacol. 2010. PMID: 20735416 Free PMC article. Review. - Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, I: Structural investigations via X-ray reflectivity from Langmuir monolayers.
Strzalka J, Liu J, Tronin A, Churbanova IY, Johansson JS, Blasie JK. Strzalka J, et al. Biophys J. 2009 May 20;96(10):4164-75. doi: 10.1016/j.bpj.2009.01.053. Biophys J. 2009. PMID: 19450487 Free PMC article. - Computational Modeling of the Nitrile Stretching Vibration of 5-Cyanoindole in Water.
Waegele MM, Gai F. Waegele MM, et al. J Phys Chem Lett. 2010 Feb 1;1(4):781-786. doi: 10.1021/jz900429z. J Phys Chem Lett. 2010. PMID: 20436926 Free PMC article.
References
- Johansson J.S. Noninactivating tandem pore domain potassium channels as attractive targets for general anesthetics. Anesth. Analg. 2003;96:1248–1250. - PubMed
- Chiara D.C., Dangott L.J., Eckenhoff R.G., Cohen J.B. Identification of nicotinic acetylcholine receptor amino acids photolabeled by the volatile anesthetic halothane. Biochemistry. 2003;42:13457–13467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources