Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I - PubMed (original) (raw)
Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I
Michaela Ulrike Gack et al. Cell Host Microbe. 2009.
Abstract
The ubiquitin ligase TRIM25 mediates Lysine 63-linked ubiquitination of the N-terminal CARD domains of the viral RNA sensor RIG-I to facilitate type I interferon (IFN) production and antiviral immunity. Here, we report that the influenza A virus nonstructural protein 1 (NS1) specifically inhibits TRIM25-mediated RIG-I CARD ubiquitination, thereby suppressing RIG-I signal transduction. A novel domain in NS1 comprising E96/E97 residues mediates its interaction with the coiled-coil domain of TRIM25, thus blocking TRIM25 multimerization and RIG-I CARD domain ubiquitination. Furthermore, a recombinant influenza A virus expressing an E96A/E97A NS1 mutant is defective in blocking TRIM25-mediated antiviral IFN response and loses virulence in mice. Our findings reveal a mechanism by which influenza virus inhibits host IFN response and also emphasize the vital role of TRIM25 in modulating antiviral defenses.
Figures
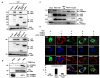
Fig. 1. Influenza A virus NS1 inhibits the TRIM25-mediated RIG-I CARD ubiquitination
(A)Whole cell lysates (WCLs) of HEK293T cells transfected with GST-RIG-I 2CARD together with vector, NS1, V5-VP35 or V5-E3L were subjected to GST-pulldown (GST-PD) followed by immunoblotting (IB) with α-GST or α-Ub. Arrows: ubiquitinated bands. (B) After transfection with RIG-I-Flag, HA-ubiquitin, and increasing amount of NS1, HEK293T were infected with SeV (50 HA units/ml) for 10 h. WCLs were subjected to immunoprecipitation (IP) with α-Flag, followed by IB with α-HA or α-Flag. (C) At 36 h post-transfection with Flag-ubiquitin, HEK293T were either mock-infected or infected with influenza A/PR/8/34 WT or ΔNS1 virus at MOI 2 for 11 h. WCLs were subjected to IP with α-RIG-I followed by IB with α-Flag or α-RIG-I. (D) WCLs of HEK293T transfected with MAVS-CARD-proline-rich domain (PRD)-Flag and GST or GST-RIG-I 2CARD together with increasing amounts of NS1 were subjected to GST-PD, followed by IB with α-Flag, α-Ub or α-GST. Arrows: ubiquitinated bands.
Fig. 2. The NS1 proteins of various influenza A virus strains interact with TRIM25
(A) HEK293T were transfected with NS1 together with vector, V5-tagged TRIM25, RING, B-boxes/CCD, SPRY (upper panels), TRIM25, B-boxes/CCD, B-boxes or CCD (lower panels). WCLs were used for IP with α-V5 followed by IB with α-NS1 or α-V5. (B) HEK293T were either mock-infected or infected with influenza A/PR/8/34 at MOI 2. At 18 h post-infection, WCLs were used for IP with α-TRIM25 or without antibody followed by IB with α-NS1 or α-TRIM25. (C) WCLs of HEK293T either mock-infected or infected with the indicated influenza A virus strains (MOI 2 for 12 h) were used for IP with α-NS1 followed by IB with α-TRIM25 or α-NS1. (D) At 20 h post-transfection with NS1 alone, TRIM25-V5 alone, NS1 together with TRIM25-V5, or NS1 together with ΔCCD-TRIM25-V5, HeLa cells were stained with α-NS1 (green), α-V5 (red) and Hoechst 33256 (nucleus, blue). More than 100 cells with expression of NS1 alone or NS1 together with TRIM25 or ΔCCD-TRIM25, respectively, were counted and percentage of the cells with cytoplasmic localization of NS1 is shown (lower left).
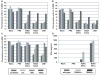
Fig. 3. Inhibition of TRIM25-mediated RIG-I signaling by WT NS1 but not E96A/E97A and R38A/K41A NS1 mutants
(A)At 48 h post-transfection of HEK293T with vector, V5-TRIM25 or TRIM25-B-boxes/CCD together with NS1 WT, R38A/K41A, or E96A/E97A, WCLs were subjected to IP with α-V5 followed by IB with α-NS1 or α-V5. (B) After transfection with GST or GST-RIG-I 2CARD together with vector, NS1 WT, R38A/K41A, or E96A/E97A, WCLs were used for GST-PD followed by IB with α-GST or α-Ub (upper two panels). WCLs were either subjected to native PAGE followed by IB with α-IRF3 (middle panel), or used for SDS-PAGE and immunoblotted with α-Phospho-IRF3 (Ser396), α-IRF3, or α-NS1 antibodies (lower panels). Arrows: ubiquitinated bands. (C) WCLs of HEK293T transfected with MAVS-CARD-PRD-Flag and GST or GST-RIG-I 2CARD together with NS1 WT or mutants were subjected to GST-PD, followed by IB with α-Flag or α-GST. Arrows: ubiquitinated bands. (D) HEK293T were transfected with GST or GST-RIG-I 2CARD together with vector or increasing amount of NS1 WT, R38A/K41A, or E96A/E97A as well as IFN-β-luciferase and pGK-β-gal. Luciferase and β-galactosidase values were determined as previously described (Gack et al., 2007). Data represent the mean + SD (n=3). (E) At 48 h post-transfection with vector, TRIM25-Flag, TRIM25-V5 and increasing amount NS1 WT, R38A/K41A, or E96A/E97A, HEK293T WCLs were subjected to IP with α-Flag followed by IB with α-V5 or α-Flag.
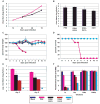
Fig. 4. Replication and pathogenicity of recombinant influenza viruses
(A) Multicycle replication of recombinant influenza viruses in A549 cells infected at an MOI of 0.001 pfu per cell. (B) Virus production from interferon-deficient 8-day old embryonated chicken eggs at 24 h post-infection with 100 pfu of each virus per egg. (C) Percent body weight loss of mice following virus infection. Female Balb/C (n = 5) were infected intranasally with 1x104 pfu of one of the indicated viruses or mock-infected with PBS. Body weights were measured for 12 consecutive days following infection. (D) Kaplan-Meier survival curves of mice infected with the indicated recombinant viruses. (E) Viral pulmonary titers of infected mice determined on day 3 and 5 post-infection (n = 3) by plaque assay. Asterisk indicates ΔNS1 not detected by plaque assay. Bars indicate standard deviation. (F) IFN bioassays measured IFN production by influenza virus-infected A549 cells. Fluorescence units represent the extent of NDV-GFP replication in Vero cells treated with supernatants from virus-infected A549 cells. Bars indicate standard deviation. The color scheme for the indicated viruses is provided at the bottom of the figure.
Fig. 5. IFN inducing/suppressing phenotypes of recombinant influenza viruses in MEFs
IFN bioassays were performed to quantitate IFN production resulting from influenza virus infection of (A) wild-type, (B) heterozygous (TRIM25+/−), or (C) homozygous (_TRIM25_−/−) MEFs. Fluorescence units represent the extent of VSV-GFP replication in L929 cells treated with supernatants from virus-infected MEF cells. The data are representative of 3 independent experiments. The legend for panels (A), (B), and (C) is provided below panel (C). (D) ELISA was performed to measure IFN-β production from wild-type, heterozygous (TRIM25+/−), or homozygous (_TRIM25_−/−) MEFs upon influenza virus infection. The results are expressed as means ± s.d. (n = replicate of 3).
Comment in
- Influenza A virus TRIMs the type I interferon response.
Ludwig S, Wolff T. Ludwig S, et al. Cell Host Microbe. 2009 May 8;5(5):420-1. doi: 10.1016/j.chom.2009.05.004. Cell Host Microbe. 2009. PMID: 19454344
Similar articles
- Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein.
Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. Rajsbaum R, et al. PLoS Pathog. 2012;8(11):e1003059. doi: 10.1371/journal.ppat.1003059. Epub 2012 Nov 29. PLoS Pathog. 2012. PMID: 23209422 Free PMC article. - Robust Lys63-Linked Ubiquitination of RIG-I Promotes Cytokine Eruption in Early Influenza B Virus Infection.
Jiang J, Li J, Fan W, Zheng W, Yu M, Chen C, Sun L, Bi Y, Ding C, Gao GF, Liu W. Jiang J, et al. J Virol. 2016 Jun 24;90(14):6263-6275. doi: 10.1128/JVI.00549-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27122586 Free PMC article. - Activation of duck RIG-I by TRIM25 is independent of anchored ubiquitin.
Miranzo-Navarro D, Magor KE. Miranzo-Navarro D, et al. PLoS One. 2014 Jan 23;9(1):e86968. doi: 10.1371/journal.pone.0086968. eCollection 2014. PLoS One. 2014. PMID: 24466302 Free PMC article. - Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I.
Oshiumi H, Matsumoto M, Seya T. Oshiumi H, et al. J Biochem. 2012 Jan;151(1):5-11. doi: 10.1093/jb/mvr111. Epub 2011 Sep 2. J Biochem. 2012. PMID: 21890623 Review. - The influenza virus NS1 protein as a therapeutic target.
Engel DA. Engel DA. Antiviral Res. 2013 Sep;99(3):409-16. doi: 10.1016/j.antiviral.2013.06.005. Epub 2013 Jun 21. Antiviral Res. 2013. PMID: 23796981 Free PMC article. Review.
Cited by
- Hypercapnia Suppresses Macrophage Antiviral Activity and Increases Mortality of Influenza A Infection via Akt1.
Casalino-Matsuda SM, Chen F, Gonzalez-Gonzalez FJ, Nair A, Dib S, Yemelyanov A, Gates KL, Budinger GRS, Beitel GJ, Sporn PHS. Casalino-Matsuda SM, et al. J Immunol. 2020 Jul 15;205(2):489-501. doi: 10.4049/jimmunol.2000085. Epub 2020 Jun 15. J Immunol. 2020. PMID: 32540997 Free PMC article. - Structural studies of the coiled-coil domain of TRIM75 reveal a tetramer architecture facilitating its E3 ligase complex.
Lou X, Ma B, Zhuang Y, Xiao X, Minze LJ, Xing J, Zhang Z, Li XC. Lou X, et al. Comput Struct Biotechnol J. 2022 Sep 5;20:4921-4929. doi: 10.1016/j.csbj.2022.08.069. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147661 Free PMC article. - Insights into pandemic respiratory viruses: manipulation of the antiviral interferon response by SARS-CoV-2 and influenza A virus.
Liu G, Gack MU. Liu G, et al. Curr Opin Immunol. 2022 Oct;78:102252. doi: 10.1016/j.coi.2022.102252. Epub 2022 Sep 14. Curr Opin Immunol. 2022. PMID: 36215931 Free PMC article. Review. - Influenza A Virus Protein PA-X Contributes to Viral Growth and Suppression of the Host Antiviral and Immune Responses.
Hayashi T, MacDonald LA, Takimoto T. Hayashi T, et al. J Virol. 2015 Jun;89(12):6442-52. doi: 10.1128/JVI.00319-15. Epub 2015 Apr 8. J Virol. 2015. PMID: 25855745 Free PMC article. - High yield production of influenza virus in Madin Darby canine kidney (MDCK) cells with stable knockdown of IRF7.
Hamamoto I, Takaku H, Tashiro M, Yamamoto N. Hamamoto I, et al. PLoS One. 2013;8(3):e59892. doi: 10.1371/journal.pone.0059892. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555825 Free PMC article.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
- Albrecht RA, Garcia-Sastre A. Suppression of innate immunity by orthomyxoviruses. In: Brasier AR, Garcia-Sastre A, Lemon SM, editors. Cellular signalling and innate immune responses to RNA virus infections. Washington: ASM Press; 2009. pp. 267–315.
- Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13:559–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 DE019085/DE/NIDCR NIH HHS/United States
- R01 AI46954/AI/NIAID NIH HHS/United States
- U19 AI062623-050003/AI/NIAID NIH HHS/United States
- R01 AI046954/AI/NIAID NIH HHS/United States
- R01 AI046954-04/AI/NIAID NIH HHS/United States
- RR00168/RR/NCRR NIH HHS/United States
- R01 AI046954-05/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- R01 CA115284/CA/NCI NIH HHS/United States
- HHNSN266200700010C/PHS HHS/United States
- U54 AI57158/AI/NIAID NIH HHS/United States
- R01 CA082057/CA/NCI NIH HHS/United States
- P51 RR000168/RR/NCRR NIH HHS/United States
- K26 RR000168/RR/NCRR NIH HHS/United States
- DE019085/DE/NIDCR NIH HHS/United States
- CA31363/CA/NCI NIH HHS/United States
- U19 AI62623/AI/NIAID NIH HHS/United States
- U19 AI083025/AI/NIAID NIH HHS/United States
- CA082057/CA/NCI NIH HHS/United States
- CA115284/CA/NCI NIH HHS/United States
- R01 CA031363/CA/NCI NIH HHS/United States
- U19 AI062623/AI/NIAID NIH HHS/United States
- HHSN266200700010C/AI/NIAID NIH HHS/United States
- U54 AI057158-05S10003/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials