Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins - PubMed (original) (raw)
Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins
Esperanza Fernández et al. Mol Syst Biol. 2009.
Abstract
The molecular complexity of mammalian proteomes demands new methods for mapping the organization of multiprotein complexes. Here, we combine mouse genetics and proteomics to characterize synapse protein complexes and interaction networks. New tandem affinity purification (TAP) tags were fused to the carboxyl terminus of PSD-95 using gene targeting in mice. Homozygous mice showed no detectable abnormalities in PSD-95 expression, subcellular localization or synaptic electrophysiological function. Analysis of multiprotein complexes purified under native conditions by mass spectrometry defined known and new interactors: 118 proteins comprising crucial functional components of synapses, including glutamate receptors, K+ channels, scaffolding and signaling proteins, were recovered. Network clustering of protein interactions generated five connected clusters, with two clusters containing all the major ionotropic glutamate receptors and one cluster with voltage-dependent K+ channels. Annotation of clusters with human disease associations revealed that multiple disorders map to the network, with a significant correlation of schizophrenia within the glutamate receptor clusters. This targeted TAP tagging strategy is generally applicable to mammalian proteomics and systems biology approaches to disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
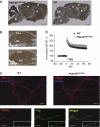
Generation of Tandem Affinity Purification (TAP)-tagged PSD-95 knockin mice. (A) Domain structure of TAP modified PSD-95. PSD-95 domains, including three PDZ (
P
SD-95/
d
iscs large/
z
ona occludens), a SH3 (
S
rc
h
omology
3
), a GK (
g
uanylate
k
inase) and C-terminal TAP-tag domain. Amino-acid sequence of the TAP tag comprising a histidine affinity tag (HAT)-domain (bold), a TEV site (underlined) and a 3XFLAG domain (bold) separated by a spacer. (B) Scheme of the targeted genomic PSD-95/Dlg4 locus. The Dlg4 allele was targeted with the TAP sequence inserted before the stop codon. Crossing the transgenic Cre-recombinase-expressing mice deleted the neomycin resistance cassette (neo) between loxP sites (bottom). Asterisk: stop codon of the coding sequence; black thick lane: TAP tag sequence; triangle: loxP site. (C) PCR genotyping of TAP-tagged PSD-95 mice, using a common forward primer PSD-95 F5 and two reverse primers PSD-95 R6 and pneoR4, which amplify the wild type (upper band) and targeted allele (lower band), respectively. (D) Immunoblot with PSD-95 antibody for immunoprecipitations. Three different heterozygous mice are shown (PSD-95TAP/+, left panel). PSD-95TAP/+ forebrain was also affinity purified with a FLAG antibody (right panel). (E) PSD-95 protein expression in wt and PSD-95TAP/TAP mouse forebrains. Brain lysates of 5, 10 and 15 μg were loaded and immunoblotted with antibodies against PSD-95 (upper panel) and tubulin (lower panel), which is a loading control. Wt, wild type; PSD-95TAP/TAP, homozygous TAP-tagged PSD-95 mice; c-, PCR water; IgG, mouse total IgG used as a negative control of the immunoprecipitation.
Figure 2
Analysis of TAP-tagged PSD-95 localization and synaptic plasticity in PSD-95TAP/TAP mice. (A) Immunohistochemical staining of PSD-95 in sagittal brain sections from PSD-95TAP/TAP and wt mice. B, brainstem; C, cortex; CB, cerebellum; H, hippocampus; S, striatum. Scale bar=1 mm. (B) Immunohistochemical staining of PSD-95 in sagittal hippocampus sections from PSD-95TAP/TAP and wt mice showing CA1, CA3 and dentate gyrus (DG). Scale bar=1 mm. (C) Synaptic localization of TAP-tagged PSD-95 in primary hippocampus neurons. DIV14 neurons from wt and PSD-95TAP/TAP mice were stained with PSD-95 and MAP2B antibodies (top panels). Three lower panel show PSD-95 and FLAG antibody staining in a culture from PSD-95TAP/TAP mice (bottom panels). Inset panels show higher magnification of synaptic puncta labeling with each antibody and merged image. Scale bar=10 μm. (D) Long-term potentiation of fEPSPs induced by theta-burst stimulation in CA1 area of hippocampal slices is similar in PSD-95TAP/TAP (13 slices from 4 animals) and wild-type mice (15 slices from 4 animals).
Figure 3
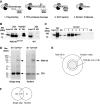
Tandem affinity purification of PSD-95 complexes. (A) Overview of the TAP protocol. In the first step, the TAP-tagged PSD-95 was captured by FLAG antibody (1) and eluted by TEV cleavage (2). Cleaved TAP-tagged PSD-95 was then captured with Ni2+–NTA–agarose beads (3) and eluted with 250 mM imidazole (4). (B) TAP-tagged PSD-95 was affinity purified with FLAG antibody from forebrain extracts from PSD-95TAP/TAP (left panel) and wt (right panel) mice, then cleaved using TEV protease (TEV) and monitored using immunoblotting with a PSD-95 antibody. The eluted (El) and column retained PSD-95 (BB) are shown. TEV protease was added to the reaction as indicated (TEV) or to control without TEV (non-TEV). Input, total lysate; El, elution after TEV reaction; BB, beads boiled with Laemmli sample buffer after TEV cleavage. (C) HAT-tagged PSD-95 purification monitored using immunoblotting against PSD-95. Following TEV cleavage the eluate (TEV El) was incubated with Ni2+–NTA–agarose, washed and eluted by imidazole 250 mM and collected in 7 fractions. TEV, TEV elution before the Ni2+ column; SN, supernatant remaining after coupling to the Ni2+ column; 1–7, fractions recovered in the imidazole elution. BB, boiled Ni2+–agarose beads after elution. (D) TAP-tagged PSD-95 complex was affinity purified using FLAG antibody (single step, left gel) and tandem (two step, right panel) from wt and PSD-95TAP/TAP forebrain and resolved by SDS–PAGE stained with colloidal Coomasie stain. The lanes were cut for mass spectrometry analysis and the identified proteins listed in Supplementary Table 1. PSD-95 and the TEV enzyme are indicated in both gels. (E) Schematic representation of the total number (301) of proteins identified in the combined single and tandem purifications. In four independent tandem purifications, a total of 158 proteins were identified and 118 appeared in at least three of four replicates (PSD-95 core complexes). (F) Venn diagram with the number of proteins from either single or tandem purifications showing the common proteins (87) and proteins masked (71) in the single-step purification.
Figure 4
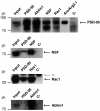
Validation of new PSD-95 interaction partners. Immunoprecipitation from forebrain extracts with indicated antibodies (labeled above panels) and immunoblotting with antibodies directed against specific proteins (labeled on the right side of each panel). Antibodies against PSD-95, Nsf, Rac1 and Ablim were used for immunoblotting. Protein molecular weight (kDa) on left. PSD-95 interaction with Arc/Arg3.1 is shown in Supplementary Figure 3B. C-, mouse total IgG was used for immunoprecipitation control; IP, antibodies used for immunoprecipitation; lc, antibody light chain.
Figure 5
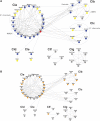
Protein interaction network of PSD-95 interacting proteins. (A) 50 proteins of the PSD-95 core complex were connected, with 119 interactions segregated into 5 clusters (Cla–Cle) forming the MCC and two separate small clusters Clf and Clg. PSD-95/Dlg4 is showed in red, primary interactors of PSD-95/Dlg4 are shown in blue and secondary interactors are shown in yellow. The glutamate receptors (NMDA, AMPA and kainate receptors) and potassium channels are bracketed. (B) Schizophrenia susceptibility genes are shown in orange.
Similar articles
- PSD-93 knock-out mice reveal that neuronal MAGUKs are not required for development or function of parallel fiber synapses in cerebellum.
McGee AW, Topinka JR, Hashimoto K, Petralia RS, Kakizawa S, Kauer FW, Aguilera-Moreno A, Wenthold RJ, Kano M, Bredt DS. McGee AW, et al. J Neurosci. 2001 May 1;21(9):3085-91. doi: 10.1523/JNEUROSCI.21-09-03085.2001. J Neurosci. 2001. PMID: 11312293 Free PMC article. - Essential contribution of the ligand-binding beta B/beta C loop of PDZ1 and PDZ2 in the regulation of postsynaptic clustering, scaffolding, and localization of postsynaptic density-95.
Nonaka M, Doi T, Fujiyoshi Y, Takemoto-Kimura S, Bito H. Nonaka M, et al. J Neurosci. 2006 Jan 18;26(3):763-74. doi: 10.1523/JNEUROSCI.2489-05.2006. J Neurosci. 2006. PMID: 16421296 Free PMC article. - Stable membrane expression of postsynaptic CaV1.2 calcium channel clusters is independent of interactions with AKAP79/150 and PDZ proteins.
Di Biase V, Obermair GJ, Szabo Z, Altier C, Sanguesa J, Bourinet E, Flucher BE. Di Biase V, et al. J Neurosci. 2008 Dec 17;28(51):13845-55. doi: 10.1523/JNEUROSCI.3213-08.2008. J Neurosci. 2008. PMID: 19091974 Free PMC article. - Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry.
Völkel P, Le Faou P, Angrand PO. Völkel P, et al. Biochem Soc Trans. 2010 Aug;38(4):883-7. doi: 10.1042/BST0380883. Biochem Soc Trans. 2010. PMID: 20658971 Review.
Cited by
- SNAP-25, a Known Presynaptic Protein with Emerging Postsynaptic Functions.
Antonucci F, Corradini I, Fossati G, Tomasoni R, Menna E, Matteoli M. Antonucci F, et al. Front Synaptic Neurosci. 2016 Mar 24;8:7. doi: 10.3389/fnsyn.2016.00007. eCollection 2016. Front Synaptic Neurosci. 2016. PMID: 27047369 Free PMC article. Review. - TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function.
Coba MP, Komiyama NH, Nithianantharajah J, Kopanitsa MV, Indersmitten T, Skene NG, Tuck EJ, Fricker DG, Elsegood KA, Stanford LE, Afinowi NO, Saksida LM, Bussey TJ, O'Dell TJ, Grant SG. Coba MP, et al. J Neurosci. 2012 Oct 3;32(40):13987-99. doi: 10.1523/JNEUROSCI.2433-12.2012. J Neurosci. 2012. PMID: 23035106 Free PMC article. - NMDA receptors are selectively partitioned into complexes and supercomplexes during synapse maturation.
Frank RA, Komiyama NH, Ryan TJ, Zhu F, O'Dell TJ, Grant SG. Frank RA, et al. Nat Commun. 2016 Apr 27;7:11264. doi: 10.1038/ncomms11264. Nat Commun. 2016. PMID: 27117477 Free PMC article. - Changes in Synaptic Proteins Precede Neurodegeneration Markers in Preclinical Alzheimer's Disease Cerebrospinal Fluid.
Lleó A, Núñez-Llaves R, Alcolea D, Chiva C, Balateu-Paños D, Colom-Cadena M, Gomez-Giro G, Muñoz L, Querol-Vilaseca M, Pegueroles J, Rami L, Lladó A, Molinuevo JL, Tainta M, Clarimón J, Spires-Jones T, Blesa R, Fortea J, Martínez-Lage P, Sánchez-Valle R, Sabidó E, Bayés À, Belbin O. Lleó A, et al. Mol Cell Proteomics. 2019 Mar;18(3):546-560. doi: 10.1074/mcp.RA118.001290. Epub 2019 Jan 3. Mol Cell Proteomics. 2019. PMID: 30606734 Free PMC article. - Dysregulated Signaling at Postsynaptic Density: A Systematic Review and Translational Appraisal for the Pathophysiology, Clinics, and Antipsychotics' Treatment of Schizophrenia.
de Bartolomeis A, Vellucci L, De Simone G, Mazza B, Barone A, Ciccarelli M. de Bartolomeis A, et al. Cells. 2023 Feb 10;12(4):574. doi: 10.3390/cells12040574. Cells. 2023. PMID: 36831241 Free PMC article. Review.
References
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A (2005) A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86: 753–758 - PubMed
- Angrand PO, Segura I, Volkel P, Ghidelli S, Terry R, Brajenovic M, Vintersten K, Klein R, Superti-Furga G, Drewes G, Kuster B, Bouwmeester T, Acker-Palmer A (2006) Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol Cell Proteomics 5: 2211–2227 - PubMed
- Bence M, Arbuckle MI, Dickson KS, Grant SG (2005) Analyses of murine postsynaptic density-95 identify novel isoforms and potential translational control elements. Brain Res Mol Brain Res 133: 143–152 - PubMed
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous