Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges - PubMed (original) (raw)
Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges
Daniel P Faith et al. Evol Bioinform Online. 2007.
Abstract
Biodiversity conservation addresses information challenges through estimations encapsulated in measures of diversity. A quantitative measure of phylogenetic diversity, "PD", has been defined as the minimum total length of all the phylogenetic branches required to span a given set of taxa on the phylogenetic tree (Faith 1992a). While a recent paper incorrectly characterizes PD as not including information about deeper phylogenetic branches, PD applications over the past decade document the proper incorporation of shared deep branches when assessing the total PD of a set of taxa. Current PD applications to macroinvertebrate taxa in streams of New South Wales, Australia illustrate the practical importance of this definition. Phylogenetic lineages, often corresponding to new, "cryptic", taxa, are restricted to a small number of stream localities. A recent case of human impact causing loss of taxa in one locality implies a higher PD value for another locality, because it now uniquely represents a deeper branch. This molecular-based phylogenetic pattern supports the use of DNA barcoding programs for biodiversity conservation planning. Here, PD assessments side-step the contentious use of barcoding-based "species" designations. Bio-informatics challenges include combining different phylogenetic evidence, optimization problems for conservation planning, and effective integration of phylogenetic information with environmental and socio-economic data.
Keywords: phylogenetic; DNA barcoding; PD; biodiversity; invertebrates; species problem.
Figures
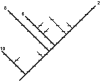
Figure 1
A hypothetical phylogenetic tree, redrawn from Faith 1992a. The path connecting those four taxa (2, 6, 8, and 10) having maximum expected feature diversity, is shown by the thickened lines. The number of tick marks traversed by this spanning path is 28, indicating the relative feature diversity for the set.
Figure 2
A phylogenetic tree example, redrawn from Faith and Williams, 2006, for taxa a through h. Taxa are found in localities p1 through p4. Taxa f, g, and h are endemic to locality p1. The PDendemism of p1 reflects the potential loss not only of the proximal connecting branches, but also the loss of the deeper branch z.
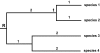
Figure 3
A figure re-drawn from Crozier et al (2005; figure 1), with species labeled 1 through 4. Crozier et al claim that the PD of species 1 and 2 is only 2 units, in counting branch lengths only back to node t. However, correct PD calculations in this comparative context would record the PD based on branches extending back to the shared root R, yielding a PD of 4 units for this set of two species.
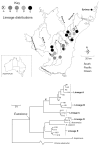
Figure 4
Phylogenetic and geographic distribution information for the “spiny crayfish” (Euastacus), as reported in Baker et al (2004) within the Sydney water supply catchment region of south-east NSW. a) The lineages labeled as A through E on the Euastacus phylogenetic tree shown in (b), are each represented only in a small number of places within the region. b) The phylogenetic pattern from Baker et al derived using the gene sequence, cytochrome c oxidase I gene (COI). Lineage A is a phylogenetic “sister” to lineage B. Given expected loss of biodiversity at localities containing lineage B, PD analysis assigns the localities containing lineage A higher priority, because the overall PD losses if both lineages were to be lost now would be high in reflecting also the loss of a shared, deeper, branch (marked X).
Similar articles
- The role of the phylogenetic diversity measure, PD, in bio-informatics: getting the definition right.
Faith DP. Faith DP. Evol Bioinform Online. 2007 Feb 19;2:277-83. Evol Bioinform Online. 2007. PMID: 19455221 Free PMC article. - Phylodiversity to inform conservation policy: An Australian example.
Laity T, Laffan SW, González-Orozco CE, Faith DP, Rosauer DF, Byrne M, Miller JT, Crayn D, Costion C, Moritz CC, Newport K. Laity T, et al. Sci Total Environ. 2015 Nov 15;534:131-43. doi: 10.1016/j.scitotenv.2015.04.113. Epub 2015 May 12. Sci Total Environ. 2015. PMID: 25976346 - Phylogenetic diversity, functional trait diversity and extinction: avoiding tipping points and worst-case losses.
Faith DP. Faith DP. Philos Trans R Soc Lond B Biol Sci. 2015 Feb 19;370(1662):20140011. doi: 10.1098/rstb.2014.0011. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25561672 Free PMC article. - Biodiversity and evolutionary history: useful extensions of the PD phylogenetic diversity assessment framework.
Faith DP. Faith DP. Ann N Y Acad Sci. 2013 Jun;1289:69-89. doi: 10.1111/nyas.12186. Ann N Y Acad Sci. 2013. PMID: 23773093 Review. - Predicting loss of evolutionary history: Where are we?
Veron S, Davies TJ, Cadotte MW, Clergeau P, Pavoine S. Veron S, et al. Biol Rev Camb Philos Soc. 2017 Feb;92(1):271-291. doi: 10.1111/brv.12228. Epub 2015 Oct 14. Biol Rev Camb Philos Soc. 2017. PMID: 26467982 Review.
Cited by
- An Overview of Bioinformatics Tools for DNA Meta-Barcoding Analysis of Microbial Communities of Bioaerosols: Digest for Microbiologists.
Mbareche H, Dumont-Leblond N, Bilodeau GJ, Duchaine C. Mbareche H, et al. Life (Basel). 2020 Sep 8;10(9):185. doi: 10.3390/life10090185. Life (Basel). 2020. PMID: 32911871 Free PMC article. Review. - Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle.
Carey HV, Walters WA, Knight R. Carey HV, et al. Am J Physiol Regul Integr Comp Physiol. 2013 Jan 1;304(1):R33-42. doi: 10.1152/ajpregu.00387.2012. Epub 2012 Nov 14. Am J Physiol Regul Integr Comp Physiol. 2013. PMID: 23152108 Free PMC article. - Intestinal Microbiota of Anser fabalis Wintering in Two Lakes in the Middle and Lower Yangtze River Floodplain.
Zhao K, Zhou D, Ge M, Zhang Y, Li W, Han Y, He G, Shi S. Zhao K, et al. Animals (Basel). 2023 Feb 17;13(4):707. doi: 10.3390/ani13040707. Animals (Basel). 2023. PMID: 36830494 Free PMC article. - The influence of timing of Maternal administration of Antibiotics during cesarean section on the intestinal Microbial colonization in Infants (MAMI-trial): study protocol for a randomised controlled trial.
Dierikx TH, Berkhout DJC, Visser L, Benninga MA, Roeselers G, de Boer NKH, de Vries JIP, de Meij TGJ. Dierikx TH, et al. Trials. 2019 Aug 5;20(1):479. doi: 10.1186/s13063-019-3552-8. Trials. 2019. PMID: 31382981 Free PMC article. - Infant Formula With a Specific Blend of Five Human Milk Oligosaccharides Drives the Gut Microbiota Development and Improves Gut Maturation Markers: A Randomized Controlled Trial.
Bosheva M, Tokodi I, Krasnow A, Pedersen HK, Lukjancenko O, Eklund AC, Grathwohl D, Sprenger N, Berger B, Cercamondi CI; 5 HMO Study Investigator Consortium. Bosheva M, et al. Front Nutr. 2022 Jul 6;9:920362. doi: 10.3389/fnut.2022.920362. eCollection 2022. Front Nutr. 2022. PMID: 35873420 Free PMC article.
References
- Andreasen K. Implications of molecular systematic analyses on the conservation of rare and threatened taxa: Contrasting examples from Malvaceae. Conserv Gen. 2005;6:399–412.
- Baker AM, Hughes JM, Dean JC, et al. Mitochondrial DNA reveals phylogenetic structuring and cryptic diversity in Australian freshwater macroinvertebrate assemblages. Mar Freshw Res. 2004;55:629–40.
- Barker GM. Phylogenetic diversity: a quantitative framework for measurement of priority and achievement in biodiversity conservation. Biol J Linn Soc. 2002;76:165–94.
- Bininda-Emonds ORP, editor. Computational Biology. XIV. Vol. 4. Springer; 2004. Phylogenetic supertrees: Combining information to reveal the Tree of Life Series; p. 550.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous