MEME SUITE: tools for motif discovery and searching - PubMed (original) (raw)
. 2009 Jul;37(Web Server issue):W202-8.
doi: 10.1093/nar/gkp335. Epub 2009 May 20.
Affiliations
- PMID: 19458158
- PMCID: PMC2703892
- DOI: 10.1093/nar/gkp335
MEME SUITE: tools for motif discovery and searching
Timothy L Bailey et al. Nucleic Acids Res. 2009 Jul.
Abstract
The MEME Suite web server provides a unified portal for online discovery and analysis of sequence motifs representing features such as DNA binding sites and protein interaction domains. The popular MEME motif discovery algorithm is now complemented by the GLAM2 algorithm which allows discovery of motifs containing gaps. Three sequence scanning algorithms--MAST, FIMO and GLAM2SCAN--allow scanning numerous DNA and protein sequence databases for motifs discovered by MEME and GLAM2. Transcription factor motifs (including those discovered using MEME) can be compared with motifs in many popular motif databases using the motif database scanning algorithm TOMTOM. Transcription factor motifs can be further analyzed for putative function by association with Gene Ontology (GO) terms using the motif-GO term association tool GOMO. MEME output now contains sequence LOGOS for each discovered motif, as well as buttons to allow motifs to be conveniently submitted to the sequence and motif database scanning algorithms (MAST, FIMO and TOMTOM), or to GOMO, for further analysis. GLAM2 output similarly contains buttons for further analysis using GLAM2SCAN and for rerunning GLAM2 with different parameters. All of the motif-based tools are now implemented as web services via Opal. Source code, binaries and a web server are freely available for noncommercial use at http://meme.nbcr.net.
Figures
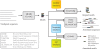
Figure 1.
Overview of the MEME Suite tools.
Figure 2.
A sample GLAM2 gapped motif.
Figure 3.
T
omtom
output. The figure shows the T
omtom
output from searching a single DNA motif against a collection of yeast transcription factor binding site motifs identified via ChIP-seq (9). T
omtom
shows that the query motif closely resembles the binding motif for transcription factor RGT1.
Figure 4.
FIMO output.
Figure 5.
GLAM2SCAN output. The figure shows the result of searching with a GLAM motif against 18 E. coli DNA sequences containing the Cyclic AMP receptor protein (CRP) binding site. Only the top 10 matches are shown.
Figure 6.
MAST output. The figure shows the result of searching with three MEME motifs against 18 E. coli DNA sequences containing the CRP binding site. The MAST output contains three representations of the results, excerpts of which are shown in the three figure panels. The _E_-value score of the overall match of the motif(s) in the input is shown in the first results section (Panel A). The second section (Panel B) displays the relative locations of significant matches of the motifs in the sequences. The third results sections gives a detailed picture of the motif matches, showing the exact location and _p_-value score of each motif match aligned above the target sequence.
Figure 7.
MEME Suite deployed with Opal (A) Opal offers versatile user access options. (B) Opal dashboard provides job history data.
Similar articles
- MEME: discovering and analyzing DNA and protein sequence motifs.
Bailey TL, Williams N, Misleh C, Li WW. Bailey TL, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W369-73. doi: 10.1093/nar/gkl198. Nucleic Acids Res. 2006. PMID: 16845028 Free PMC article. - MEME-ChIP: motif analysis of large DNA datasets.
Machanick P, Bailey TL. Machanick P, et al. Bioinformatics. 2011 Jun 15;27(12):1696-7. doi: 10.1093/bioinformatics/btr189. Epub 2011 Apr 12. Bioinformatics. 2011. PMID: 21486936 Free PMC article. - FIMO: scanning for occurrences of a given motif.
Grant CE, Bailey TL, Noble WS. Grant CE, et al. Bioinformatics. 2011 Apr 1;27(7):1017-8. doi: 10.1093/bioinformatics/btr064. Epub 2011 Feb 16. Bioinformatics. 2011. PMID: 21330290 Free PMC article. - Discovering sequence motifs.
Bailey TL. Bailey TL. Methods Mol Biol. 2008;452:231-51. doi: 10.1007/978-1-60327-159-2_12. Methods Mol Biol. 2008. PMID: 18566768 Review. - A Review on Planted (l, d) Motif Discovery Algorithms for Medical Diagnose.
Mohanty S, Pattnaik PK, Al-Absi AA, Kang DK. Mohanty S, et al. Sensors (Basel). 2022 Feb 5;22(3):1204. doi: 10.3390/s22031204. Sensors (Basel). 2022. PMID: 35161949 Free PMC article. Review.
Cited by
- Role of Squalene Epoxidase Gene (SQE1) in the Response of the Lichen Lobaria pulmonaria to Temperature Stress.
Onele AO, Swid MA, Leksin IY, Rakhmatullina DF, Galeeva EI, Beckett RP, Minibayeva FV, Valitova JN. Onele AO, et al. J Fungi (Basel). 2024 Oct 9;10(10):705. doi: 10.3390/jof10100705. J Fungi (Basel). 2024. PMID: 39452657 Free PMC article. - Genome-wide analysis of the salmonella Fis regulon and its regulatory mechanism on pathogenicity islands.
Wang H, Liu B, Wang Q, Wang L. Wang H, et al. PLoS One. 2013 May 23;8(5):e64688. doi: 10.1371/journal.pone.0064688. Print 2013. PLoS One. 2013. PMID: 23717649 Free PMC article. - Genome-Wide Identification of Tannase Genes and Their Function of Wound Response and Astringent Substances Accumulation in Juglandaceae.
Wang J, Wang K, Lyu S, Huang J, Huang C, Xing Y, Wang Y, Xu Y, Li P, Hong J, Xi J, Si X, Ye H, Li Y. Wang J, et al. Front Plant Sci. 2021 May 17;12:664470. doi: 10.3389/fpls.2021.664470. eCollection 2021. Front Plant Sci. 2021. PMID: 34079571 Free PMC article. - Runx2 and Runx3 differentially regulate articular chondrocytes during surgically induced osteoarthritis development.
Nagata K, Hojo H, Chang SH, Okada H, Yano F, Chijimatsu R, Omata Y, Mori D, Makii Y, Kawata M, Kaneko T, Iwanaga Y, Nakamoto H, Maenohara Y, Tachibana N, Ishikura H, Higuchi J, Taniguchi Y, Ohba S, Chung UI, Tanaka S, Saito T. Nagata K, et al. Nat Commun. 2022 Oct 19;13(1):6187. doi: 10.1038/s41467-022-33744-5. Nat Commun. 2022. PMID: 36261443 Free PMC article. - Genome-wide prediction of activating regulatory elements in rice by combining STARR-seq with FACS.
Tian W, Huang X, Ouyang X. Tian W, et al. Plant Biotechnol J. 2022 Dec;20(12):2284-2297. doi: 10.1111/pbi.13907. Epub 2022 Aug 26. Plant Biotechnol J. 2022. PMID: 36028476 Free PMC article.
References
- Bailey TL, Elkan CP. Fitting a mixture model by expectation-maximization to discover motifs in biopolymers. In: Altman R, Brutlag D, Karp P, Lathrop R, Searls D, editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. - PubMed
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases