Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs - PubMed (original) (raw)
Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs
John R Collette et al. Mol Biol Cell. 2009 Jul.
Abstract
Clathrin is involved in vesicle formation in the trans-Golgi network (TGN)/endosomal system and during endocytosis. Clathrin recruitment to membranes is mediated by the clathrin heavy chain (HC) N-terminal domain (TD), which forms a seven-bladed beta-propeller. TD binds membrane-associated adaptors, which have short peptide motifs, either the clathrin-box (CBM) and/or the W-box; however, the importance of the TD binding sites for these motifs has not been tested in vivo. We investigated the importance of the TD in clathrin function by generating 1) mutations in the yeast HC gene (CHC1) to disrupt the binding sites for the CBM and W-box (chc1-box), and 2) four TD-specific temperature-sensitive alleles of CHC1. We found that TD is important for the retention of resident TGN enzymes and endocytosis of alpha-factor; however, the known adaptor binding sites are not necessary, because chc1-box caused little to no effect on trafficking pathways involving clathrin. The Chc1-box TD was able to interact with the endocytic adaptor Ent2 in a CBM-dependent manner, and HCs encoded by chc1-box formed clathrin-coated vesicles. These data suggest that additional or alternative binding sites exist on the TD propeller to help facilitate the recruitment of clathrin to sites of vesicle formation.
Figures
Figure 1.
Structure of the clathrin heavy chain TD. (A) A homology model of the yeast TD (green) with blades 1–7 of the β-propeller numbered in bold. Side chains from amino acid residues that were mutated in the chc1-TD-ts alleles and the chc1-box allele are shown and color coded for each allele as follows: chc1-1 and chc1-3 (dark blue), chc1-2 (red), chc1-4 (yellow), and chc1-box (purple). (B) Sequence alignment of the yeast TD (residues 1-359) and human TD (residues 1-353) regions of clathrin heavy chain. Residues that were mutated in the chc1-TD-ts alleles and the chc1-box allele are highlighted according to the same color scheme used in A. An asterisk (*) denotes identical residues between the yeast and human sequence, a colon (:) denotes a conserved substitution between sequences, and a period (.) denotes a semiconserved substitution between sequences. (C) Alignment of the seven blades of the TD β-propeller structure. The approximate boundaries for every strand in each blade are denoted above the alignment. θ denotes conserved hydrophobic residues in both the yeast and human sequence. The S75P mutation of chc1-1 is also present in chc1-3.
Figure 2.
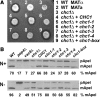
The chc1-box mutant is not temperature-sensitive for growth. (A) SL82 (_chc1_Δ + pAP4[CEN, _CHC1_]), SL5177 (_chc1_Δ + pJRC2[CEN, _chc1-1_]), SL5171 (_chc1_Δ + pJRC3[CEN, _chc1-2_]), SL5181 (_chc1_Δ + pJRC4[CEN, _chc1-3_]), SL5178 (_chc1_Δ + pJRC5[CEN, _chc1-4_]), and SL5634 (_chc1_Δ + pJRC19[CEN, _chc1-box_]) cells were diluted to 107 cells/ml, and fourfold serial dilutions were spotted onto plates and grown at the indicated temperatures for 3 d. (B) Immunoblots of extracts from _chc1_Δ yeast (SL12) transformed with pUN30 (CEN, empty), pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box) were detected with anti-Chc1 antibodies. Cells were grown continuously at 25°C, and then one-half of each sample was removed and incubated for 90 min at 37°C before harvesting. To verify equal protein loading, blots also were probed with antibodies to tubulin.
Figure 3.
chc1-TD-ts alleles cause TGN sorting and Ape1 processing defects. (A) Halo assays for _MAT_α _chc1_Δ yeast (SL119) transformed with pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), pSL6-box (CEN, chc1-box), and wild-type controls, SL1462 (MATa) and SL1463, (_MAT_α) spotted onto a lawn of MATa (BJ3556) and grown at 30°C for 2 d. A zone of growth inhibition demonstrates secretion of mature α-factor. (B) Ape1 processing: immunoblots of wild-type (SL1463), _pep4_Δ (BJ3502), _vac8_Δ (SL2567), _chc1_Δ (SL12), and SL12 transformed with pUN30 (CEN, empty), pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pSL6-box (CEN, chc1-box), grown in normal medium (N+) or nitrogen starvation medium (N−) at 30°C and blotted with anti-Ape1 antibodies. The amounts of precursor (pApe1) and mature aminopeptidase (mApe1) were determined by densitometry, and the percentage of mApe1 is reported relative to total mApe1 + pApe1 for each.
Figure 4.
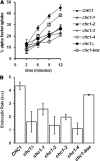
The chc1-box allele, but not chc1-TD-ts alleles, rescues the endocytic phenotype of _chc1_Δ. MATa _chc1_Δ bar1-1 (SL3593) was transformed with pUN30 (CEN, empty), pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box). α-factor internalization was analyzed at 37°C as described in Materials and Methods.
Figure 5.
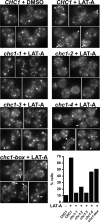
GFP-Clc1p accumulation at the cell cortex in chc1-TD alleles in the presence of LAT-A. _GFP-CLC1 ABP1-mRFP chc1_Δ (SL5538) was transformed with pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box). Cells grown to log phase at 25°C were treated with LAT-A (250 μM) for 20 min at 25°C before imaging. Abp1-mRFP was completely cytosolic after 20 min in LAT-A, indicating the disassembly of actin (data not shown). Arrows indicate the accumulation of clathrin at the cell cortex. Cells that showed strong cortical puncta or rim GFP staining were considered to have accumulated clathrin. The percentage of cells for each condition that contained cortical GFP-Clc1 accumulation is shown in the graph (n = 100–150).
Figure 6.
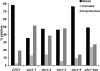
TIRFM of clathrin patch dynamics at the cell cortex Graph show percentages of GFP-Clc1 patch types observed in CHC1 and chc1-TD mutants at 25°C. Between 50 and 80 GFP-Clc1 patches for each strain were defined as mobile, immobile or unproductive as described in Materials and Methods. Strains are the same as in Figure 5.
Figure 7.
HC-TD mutations disrupt interactions with clathrin adaptors Ent1, Ent2, and Apl2. (A) A two-hybrid reporter strain (SL2793) was transformed with either the empty GAL4 binding domain (GBD) plasmid pGBDU-C1 or pGBD-ENT1 in combination with either the empty GAL4 activation domain (GAD) plasmid pGAD-C1 or pGAD-CHC1-TD(1-363). Cells from each set of transformants were diluted to 5 × 106 cells/ml, spotted onto C-LEU-URA (total growth) and C-LEU-URA-ADE (activation of the GAL2:ADE2 reporter) plates, and grown at 30°C for 3 d. (B) SL2793 was cotransformed with pGBD-ENT1 and each of the following GAD-CHC1-TD plasmids: pJRC8 (GAD-CHC1-TD), pJRC14 (GAD-CHC1-TD[_chc1-1_]), pJRC15 (GAD-CHC1-TD[_chc1-2_]), pJRC16 (pGAD-CHC1-TD[_chc1-3_]), pJRC17 (GAD-CHC1-TD[_chc1-4_]), or pJRC18 (GAD-CHC1-TD[_chc1-box_]) and plated and grown as described in A. (C) PJ694α was transformed with pGBDU-C1 or the following plasmids: pJRC6 (GBD-CHC1-TD), pJRC20 (GBD-CHC1-TD[_chc1-1_]), pJRC21 (GBD-CHC1-TD[_chc1-2_]), pJRC22 (GBD-CHC1-TD[_chc1-3_]), pJRC23 (GBD-CHC1-TD[_chc1-4_]), pJRC24 (GBD-CHC1-TD[_chc1-box_]) and mated with PJ694a transformed with either the empty GAD plasmid pOAD, pOAD-Ent2, pOAD-Ent2(ΔCBM), or pOAD-Apl2. Diploids were selected on C-LEU-URA plates, and then activation of the ADE2 reporter was assayed as described above.
Figure 8.
Clathrin HC encoded by the chc1-box allele forms coated vesicles. Clathrin-coated vesicles were prepared from _chc1_Δ cells (SL12) transformed with pAP4 (CEN, CHC1) or pJRC19 (CEN, chc1-box), grown at 30°C, and fractionated on a Sephacryl S-1000 column. Column fractions 22–48 for each preparation were immunoblotted with anti-Chc1 and anti-Apm1 antibodies.
Figure 9.
The chc1-box allele restores calcofluor resistance to _chs6_Δ _chc1_Δ (Δ
Δ) cells. (A) SL119 (_chc1_Δ), YRV19 (_chs6_Δ), SL5719 (_chs6_Δ _chc1_Δ[ΔΔ]), and ΔΔ + pAP4 (CEN, CHC1) cells were diluted to 1 × 107 cells/ml, and fourfold serial dilutions were plated onto both YEPD plates and YEPD plates containing 50 μg/ml calcofluor white and incubated at 30°C for 3–5 d. (B) ΔΔ cells transformed with pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box) were diluted to 1 × 107 cells/ml, and fourfold serial dilutions were plated as described in A and incubated at 30°C (or 25°C where indicated).
Similar articles
- Nuclear Magnetic Resonance Structural Mapping Reveals Promiscuous Interactions between Clathrin-Box Motif Sequences and the N-Terminal Domain of the Clathrin Heavy Chain.
Zhuo Y, Cano KE, Wang L, Ilangovan U, Hinck AP, Sousa R, Lafer EM. Zhuo Y, et al. Biochemistry. 2015 Apr 28;54(16):2571-80. doi: 10.1021/acs.biochem.5b00065. Epub 2015 Apr 16. Biochemistry. 2015. PMID: 25844500 Free PMC article. - In vivo andin vitro studies of adaptor-clathrin interaction.
Feliciano D, Bultema JJ, Ambrosio AL, Di Pietro SM. Feliciano D, et al. J Vis Exp. 2011 Jan 26;(47):2352. doi: 10.3791/2352. J Vis Exp. 2011. PMID: 21307828 Free PMC article. - Creating a chimeric clathrin heavy chain that functions independently of yeast clathrin light chain.
Boettner DR, Segarra VA, Moorthy BT, de León N, Creagh J, Collette JR, Malhotra A, Lemmon SK. Boettner DR, et al. Traffic. 2016 Jul;17(7):754-68. doi: 10.1111/tra.12401. Epub 2016 May 11. Traffic. 2016. PMID: 27062026 Free PMC article. - Getting in touch with the clathrin terminal domain.
Lemmon SK, Traub LM. Lemmon SK, et al. Traffic. 2012 Apr;13(4):511-9. doi: 10.1111/j.1600-0854.2011.01321.x. Epub 2012 Jan 13. Traffic. 2012. PMID: 22239657 Free PMC article. Review. - Vesicle formation at the plasma membrane and trans-Golgi network: the same but different.
McNiven MA, Thompson HM. McNiven MA, et al. Science. 2006 Sep 15;313(5793):1591-4. doi: 10.1126/science.1118133. Science. 2006. PMID: 16973870 Review.
Cited by
- Evolving models for assembling and shaping clathrin-coated pits.
Chen Z, Schmid SL. Chen Z, et al. J Cell Biol. 2020 Sep 7;219(9):e202005126. doi: 10.1083/jcb.202005126. J Cell Biol. 2020. PMID: 32770195 Free PMC article. Review. - Functional analysis of interaction sites on the N-terminal domain of clathrin heavy chain.
Willox AK, Royle SJ. Willox AK, et al. Traffic. 2012 Jan;13(1):70-81. doi: 10.1111/j.1600-0854.2011.01289.x. Epub 2011 Oct 20. Traffic. 2012. PMID: 21939487 Free PMC article. - Activation of the Rab7 GTPase by the MON1-CCZ1 Complex Is Essential for PVC-to-Vacuole Trafficking and Plant Growth in Arabidopsis.
Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L. Cui Y, et al. Plant Cell. 2014 May;26(5):2080-2097. doi: 10.1105/tpc.114.123141. Epub 2014 May 13. Plant Cell. 2014. PMID: 24824487 Free PMC article. - Clathrin binding by the adaptor Ent5 promotes late stages of clathrin coat maturation.
Hung CW, Duncan MC. Hung CW, et al. Mol Biol Cell. 2016 Apr 1;27(7):1143-53. doi: 10.1091/mbc.E15-08-0588. Epub 2016 Feb 3. Mol Biol Cell. 2016. PMID: 26842894 Free PMC article. - Nuclear Magnetic Resonance Structural Mapping Reveals Promiscuous Interactions between Clathrin-Box Motif Sequences and the N-Terminal Domain of the Clathrin Heavy Chain.
Zhuo Y, Cano KE, Wang L, Ilangovan U, Hinck AP, Sousa R, Lafer EM. Zhuo Y, et al. Biochemistry. 2015 Apr 28;54(16):2571-80. doi: 10.1021/acs.biochem.5b00065. Epub 2015 Apr 16. Biochemistry. 2015. PMID: 25844500 Free PMC article.
References
- Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C., Wakeham D. E. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. - PubMed
- Bumbulis M. J., Wroblewski G., McKean D., Setzer D. R. Genetic analysis of Xenopus transcription factor IIIA. J. Mol. Biol. 1998;284:1307–1322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32-GM084677/GM/NIGMS NIH HHS/United States
- T32-HL07188/HL/NHLBI NIH HHS/United States
- F32 GM084677/GM/NIGMS NIH HHS/United States
- R01 DK053249/DK/NIDDK NIH HHS/United States
- R01 GM077349-02/GM/NIGMS NIH HHS/United States
- GM-084677/GM/NIGMS NIH HHS/United States
- T32 HL007188/HL/NHLBI NIH HHS/United States
- R01 GM055796/GM/NIGMS NIH HHS/United States
- R01 GM077349/GM/NIGMS NIH HHS/United States
- R01 GM055796-12/GM/NIGMS NIH HHS/United States
- DK-53249/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous