High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia - PubMed (original) (raw)
. 2009 Jul 16;114(3):647-50.
doi: 10.1182/blood-2009-02-206722. Epub 2009 May 20.
Takaomi Sanda, Ruta Grebliunaite, Arkaitz Carracedo, Leonardo Salmena, Yebin Ahn, Suzanne Dahlberg, Donna Neuberg, Lisa A Moreau, Stuart S Winter, Richard Larson, Jianhua Zhang, Alexei Protopopov, Lynda Chin, Pier Paolo Pandolfi, Lewis B Silverman, Stephen P Hunger, Stephen E Sallan, A Thomas Look
Affiliations
- PMID: 19458356
- PMCID: PMC2713461
- DOI: 10.1182/blood-2009-02-206722
High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia
Alejandro Gutierrez et al. Blood. 2009.
Abstract
To more comprehensively assess the pathogenic contribution of the PTEN-PI3K-AKT pathway to T-cell acute lymphoblastic leukemia (T-ALL), we examined diagnostic DNA samples from children with T-ALL using array comparative genomic hybridization and sequence analysis. Alterations of PTEN, PI3K, or AKT were identified in 47.7% of 44 cases. There was a striking clustering of PTEN mutations in exon 7 in 12 cases, all of which were predicted to truncate the C2 domain without disrupting the phosphatase domain of PTEN. Induction chemotherapy failed to induce remission in 3 of the 4 patients whose lymphoblasts harbored PTEN deletions at the time of diagnosis, compared with none of the 12 patients with mutations of PTEN exon 7 (P = .007), suggesting that PTEN deletion has more adverse therapeutic consequences than mutational disruptions that preserve the phosphatase domain. These findings add significant support to the rationale for the development of therapies targeting the PTEN-PI3K-AKT pathway in T-ALL.
Figures
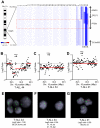
Figure 1
PTEN deletions in T-ALL. (A) Array CGH was performed with genomic DNA from diagnostic specimens collected from 47 children with T-ALL. The data are shown as a dChip plot of CGH segmented log2 copy number ratios at the PTEN locus. The red box denotes the location of the PTEN coding sequence. White arrowheads point to cases with segmented log2 copy number ratios of less than −0.5 involving the PTEN coding sequence. Two samples on which CGH was unsuccessful (T-ALL 36 and 37) were excluded from analysis. (B-D) Raw CGH data from representative patient samples. Red lines represent the segmented log2 copy number ratio shown in panel A. (E-G) FISH analysis of representative cases confirmed the deletions identified by CGH. Orange, PTEN probe; green, centromere 10 probe. Images were obtained with an Axio Imager A1 fluorescence microscope (Carl Zeiss) using a 100× Alpha Apochromatic Plan oil-immersion objective (Carl Zeiss), a JAI CV-M4+CL progressive scan camera (JAI Inc), and Genus acquisition software version 3.92 build 7 (Genetix USA). Note that the Genus cytogenetic image acquisition software applies an automated “thresholding” algorithm that sets a signal intensity threshold below which any signal is considered background and thus excluded from the final composite image. During image acquisition, all images generated by the software were compared to the view from the microscope to confirm that they were fully representative. (B,E) Homozygous deletions had log2 ratios of −1.26 (case 44) and −4.11 (case 45). Cells were available for FISH on case 44 and clearly showed homozygous loss of PTEN. (C,F) The CGH detection of a heterozygous deletion in case 34 (log2 ratio, −0.54) correlated with the detection of PTEN deletion by FISH on 1 allele in 21% of the cells examined. (D,G) Case 21 retained both PTEN alleles intact by FISH and CGH.
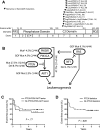
Figure 2
Mutations of PTEN and the PI3K-AKT pathway in T-ALL. (A) Sequencing of PTEN in 44 of the primary samples shown in Figure 1 identified nonsynonymous sequence alterations in 12 of these samples, all of which were predicted to disrupt the PTEN protein within an 18-amino acid region of the C2 domain. Note that the specific mutations in cases 14 and 27 were impossible to determine because of the presence of 2 simultaneous frameshift sequences. (B) Targeted sequencing of PIK3R1, PIK3CA, and AKT1-3 exons known to be mutated in human cancer identified nonsynonymous sequence alterations in PTEN and the PI3K-AKT pathway in 47.7% of primary T-ALL cases. Lesions within the PTEN-PI3K-AKT pathway were mutually exclusive. Abnormalities in the NF1 and RAS genes were also identified but were not solely associated with PTEN-PI3K-AKT pathway abnormalities. *Novel in-frame insertion/deletions. (C-D) Kaplan-Meier event-free survival curves for the 44 cases analyzed by CGH and sequencing demonstrate that, overall, genetic alterations of the PTEN-PI3K-AKT pathway did not predict event-free survival, whereas deletions of PTEN were significantly associated with early treatment failure.
Similar articles
- The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia.
Zuurbier L, Petricoin EF 3rd, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines JG, Smits WK, Sonneveld E, Veerman AJ, Kamps WA, Horstmann M, Pieters R, Meijerink JP. Zuurbier L, et al. Haematologica. 2012 Sep;97(9):1405-13. doi: 10.3324/haematol.2011.059030. Epub 2012 Apr 4. Haematologica. 2012. PMID: 22491738 Free PMC article. - The relevance of PTEN-AKT in relation to NOTCH1-directed treatment strategies in T-cell acute lymphoblastic leukemia.
Mendes RD, Canté-Barrett K, Pieters R, Meijerink JP. Mendes RD, et al. Haematologica. 2016 Sep;101(9):1010-7. doi: 10.3324/haematol.2016.146381. Haematologica. 2016. PMID: 27582570 Free PMC article. Review. - Single-cell profiling of pediatric T-cell acute lymphoblastic leukemia: Impact of PTEN exon 7 mutation on PI3K/Akt and JAK-STAT signaling pathways.
Bonaccorso P, Bugarin C, Buracchi C, Fazio G, Biondi A, Lo Nigro L, Gaipa G. Bonaccorso P, et al. Cytometry B Clin Cytom. 2020 Nov;98(6):491-503. doi: 10.1002/cyto.b.21882. Epub 2020 Jun 1. Cytometry B Clin Cytom. 2020. PMID: 32479694 - PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability.
Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, Nowill AE, Leslie NR, Cardoso AA, Barata JT. Silva A, et al. J Clin Invest. 2008 Nov;118(11):3762-74. doi: 10.1172/JCI34616. Epub 2008 Oct 1. J Clin Invest. 2008. PMID: 18830414 Free PMC article. - New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia.
Hales EC, Taub JW, Matherly LH. Hales EC, et al. Cell Signal. 2014 Jan;26(1):149-61. doi: 10.1016/j.cellsig.2013.09.021. Epub 2013 Oct 16. Cell Signal. 2014. PMID: 24140475 Review.
Cited by
- Wnt signaling mediates oncogenic synergy between Akt and Dlx5 in T-cell lymphomagenesis by enhancing cholesterol synthesis.
Tan Y, Sementino E, Liu Z, Cai KQ, Testa JR. Tan Y, et al. Sci Rep. 2020 Sep 28;10(1):15837. doi: 10.1038/s41598-020-72822-w. Sci Rep. 2020. PMID: 32985581 Free PMC article. - Phase I study of UCN-01 and perifosine in patients with relapsed and refractory acute leukemias and high-risk myelodysplastic syndrome.
Gojo I, Perl A, Luger S, Baer MR, Norsworthy KJ, Bauer KS, Tidwell M, Fleckinger S, Carroll M, Sausville EA. Gojo I, et al. Invest New Drugs. 2013 Oct;31(5):1217-27. doi: 10.1007/s10637-013-9937-8. Epub 2013 Feb 27. Invest New Drugs. 2013. PMID: 23443507 Free PMC article. Clinical Trial. - Zebrafish as a model for leukemia and other hematopoietic disorders.
Rasighaemi P, Basheer F, Liongue C, Ward AC. Rasighaemi P, et al. J Hematol Oncol. 2015 Mar 28;8:29. doi: 10.1186/s13045-015-0126-4. J Hematol Oncol. 2015. PMID: 25884214 Free PMC article. Review. - Simultaneous inhibition of pan-phosphatidylinositol-3-kinases and MEK as a potential therapeutic strategy in peripheral T-cell lymphomas.
Martín-Sánchez E, Rodríguez-Pinilla SM, Sánchez-Beato M, Lombardía L, Domínguez-González B, Romero D, Odqvist L, García-Sanz P, Wozniak MB, Kurz G, Blanco-Aparicio C, Mollejo M, Alves FJ, Menárguez J, González-Palacios F, Rodríguez-Peralto JL, Ortiz-Romero PL, García JF, Bischoff JR, Piris MA. Martín-Sánchez E, et al. Haematologica. 2013 Jan;98(1):57-64. doi: 10.3324/haematol.2012.068510. Epub 2012 Jul 16. Haematologica. 2013. PMID: 22801959 Free PMC article. - The Superior Cytotoxicity of Dual Targeting of BCR/ABL and PI3K in K562 Cells: Proposing a Novel Therapeutic Potential for the Treatment of CML.
Shiri Heris R, Pourbagheri-Sigaroodi A, Yousefi AM, Bashash D. Shiri Heris R, et al. Indian J Hematol Blood Transfus. 2022 Jan;38(1):51-60. doi: 10.1007/s12288-021-01434-9. Epub 2021 Apr 1. Indian J Hematol Blood Transfus. 2022. PMID: 35125711 Free PMC article.
References
- Goldberg JM, Silverman LB, Levy DE, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–3622. - PubMed
- Bassan R, Gatta G, Tondini C, Willemze R. Adult acute lymphoblastic leukaemia. Crit Rev Oncol Hematol. 2004;50:223–261. - PubMed
- Einsiedel HG, von Stackelberg A, Hartmann R, et al. Long-term outcome in children with relapsed ALL by risk-stratified salvage therapy: results of trial acute lymphoblastic leukemia-relapse study of the Berlin-Frankfurt-Munster Group 87. J Clin Oncol. 2005;23:7942–7950. - PubMed
- Chow LM, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241:184–196. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous