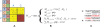SHRiMP: accurate mapping of short color-space reads - PubMed (original) (raw)
SHRiMP: accurate mapping of short color-space reads
Stephen M Rumble et al. PLoS Comput Biol. 2009 May.
Abstract
The development of Next Generation Sequencing technologies, capable of sequencing hundreds of millions of short reads (25-70 bp each) in a single run, is opening the door to population genomic studies of non-model species. In this paper we present SHRiMP - the SHort Read Mapping Package: a set of algorithms and methods to map short reads to a genome, even in the presence of a large amount of polymorphism. Our method is based upon a fast read mapping technique, separate thorough alignment methods for regular letter-space as well as AB SOLiD (color-space) reads, and a statistical model for false positive hits. We use SHRiMP to map reads from a newly sequenced Ciona savignyi individual to the reference genome. We demonstrate that SHRiMP can accurately map reads to this highly polymorphic genome, while confirming high heterozygosity of C. savignyi in this second individual. SHRiMP is freely available at http://compbio.cs.toronto.edu/shrimp.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Data flow and processing within the SHRiMP.
Candidate mapping locations are first discovered by the seed scanner and then validated by the vectorized Smith-Waterman algorithm, computing only a score. Top scoring hits are then fully aligned by a platform-specific algorithm (i.e. letter-space for Solexa data and color-space for SOLiD data). Statistical confidence for the final mappings are then computed using the PROBCALC utility.
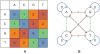
Figure 2. Two representations of the color-space (dibase) encoding used by the AB SOLiD sequencing system.
A: The standard representation, with the first and second letter of the queried pair along the horizontal and vertical axes, respectively. B: The equivalent Finite State Automaton representation, with edges labelled with the readouts and nodes corresponding to the basepairs of the underlying genome.
Figure 3. Various mutation and error events, and their effects on the color-code readouts.
The reference genome is labeled G and the read R. A: A perfect alignment; B: In case of a sequencing error (the 2 should have been read as a 0) the rest of the read no longer matches the genome in letter-space; C: In case of a SNP two adjacent colors do not match the genome, but all subsequent letters do match. However, D: only 3 of the 9 possible color changes represent valid SNPs; E: the rules for deciding which insertion and deletion events are valid are even more complex, as indels can also change adjacent color readouts.
Figure 4. Color-space (dibase) sequence alignment.
A: The Dynamic Programming (DP) representation, B: recurrences, and C: alignment of a letter space sequence to a color-space read with a sequencing error. Within the DP matrix we simultaneously align all of the four possible translations (vertical) to the reference genome (horizontal); however the alignment can transition between translations by paying the crossover penalty. This is illustrated by the fourth recurrence, where the third index ( ) corresponds to the translation currently being used. In the alignment (C) after the sequencing error, the original translation of the read (starting from a T) no longer matches, but a different one (starting from a C) does.
) corresponds to the translation currently being used. In the alignment (C) after the sequencing error, the original translation of the read (starting from a T) no longer matches, but a different one (starting from a C) does.
Figure 5. Size distribution of indels.
(A) and distance between adjacent SNPs (B) detected by SHRiMP. The distance between adjacent SNPs shows a clear 3-periodicity, due to the fact that a significant fraction of the non-repetitive C. savignyi genome is coding.
Figure 6. SHRiMP Hashing technique & Vectorized Alignment algorithm.
A: Overview of the k-mer filtering stage within SHRiMP: A window is moved along the genome. If a particular read has a preset number of k-mers within the window the vectorized Smith-Waterman stage is run to align the read to the genome. B: Schematic of the vectorized-implementation of the Needleman-Wunsch algorithm. The red cells are the vector being computed, on the basis of the vectors computed in the last step (yellow) and the next-to-last (blue). The match/mismatch vector for the diagonal is determined by comparing one sequence with the other one reversed (indicated by the red arrow below). To obtain the set of match/mismatch positions for the next diagonal, the lower sequence needs to be shifted to the right.
Similar articles
- SHRiMP2: sensitive yet practical SHort Read Mapping.
David M, Dzamba M, Lister D, Ilie L, Brudno M. David M, et al. Bioinformatics. 2011 Apr 1;27(7):1011-2. doi: 10.1093/bioinformatics/btr046. Epub 2011 Jan 28. Bioinformatics. 2011. PMID: 21278192 - Fast and accurate short read alignment with Burrows-Wheeler transform.
Li H, Durbin R. Li H, et al. Bioinformatics. 2009 Jul 15;25(14):1754-60. doi: 10.1093/bioinformatics/btp324. Epub 2009 May 18. Bioinformatics. 2009. PMID: 19451168 Free PMC article. - ComB: SNP calling and mapping analysis for color and nucleotide space platforms.
Souaiaia T, Frazier Z, Chen T. Souaiaia T, et al. J Comput Biol. 2011 Jun;18(6):795-807. doi: 10.1089/cmb.2011.0027. Epub 2011 May 12. J Comput Biol. 2011. PMID: 21563978 Free PMC article. - A survey of sequence alignment algorithms for next-generation sequencing.
Li H, Homer N. Li H, et al. Brief Bioinform. 2010 Sep;11(5):473-83. doi: 10.1093/bib/bbq015. Epub 2010 May 11. Brief Bioinform. 2010. PMID: 20460430 Free PMC article. Review. - Mapping RNA-seq Reads with STAR.
Dobin A, Gingeras TR. Dobin A, et al. Curr Protoc Bioinformatics. 2015 Sep 3;51:11.14.1-11.14.19. doi: 10.1002/0471250953.bi1114s51. Curr Protoc Bioinformatics. 2015. PMID: 26334920 Free PMC article. Review.
Cited by
- Fine de novo sequencing of a fungal genome using only SOLiD short read data: verification on Aspergillus oryzae RIB40.
Umemura M, Koyama Y, Takeda I, Hagiwara H, Ikegami T, Koike H, Machida M. Umemura M, et al. PLoS One. 2013 May 7;8(5):e63673. doi: 10.1371/journal.pone.0063673. Print 2013. PLoS One. 2013. PMID: 23667655 Free PMC article. - Web-based bioinformatics workflows for end-to-end RNA-seq data computation and analysis in agricultural animal species.
Li W, Richter RA, Jung Y, Zhu Q, Li RW. Li W, et al. BMC Genomics. 2016 Sep 27;17(1):761. doi: 10.1186/s12864-016-3118-z. BMC Genomics. 2016. PMID: 27678198 Free PMC article. - Repeat infections with chlamydia in women may be more transcriptionally active with lower responses from some immune genes.
Huston WM, Lawrence A, Wee BA, Thomas M, Timms P, Vodstrcil LA, McNulty A, McIvor R, Worthington K, Donovan B, Phillips S, Chen MY, Fairley CK, Hocking JS. Huston WM, et al. Front Public Health. 2022 Oct 10;10:1012835. doi: 10.3389/fpubh.2022.1012835. eCollection 2022. Front Public Health. 2022. PMID: 36299763 Free PMC article. - Comparative genome analysis of Lactobacillus casei strains isolated from Actimel and Yakult products reveals marked similarities and points to a common origin.
Douillard FP, Kant R, Ritari J, Paulin L, Palva A, de Vos WM. Douillard FP, et al. Microb Biotechnol. 2013 Sep;6(5):576-87. doi: 10.1111/1751-7915.12062. Epub 2013 Jul 1. Microb Biotechnol. 2013. PMID: 23815335 Free PMC article.
References
- Bowtie. http://bowtie-bio.sourceforge.net.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources