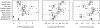Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples - PubMed (original) (raw)
Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples
Balazs Gyorffy et al. PLoS One. 2009.
Abstract
Background: Several preprocessing algorithms for Affymetrix gene expression microarrays have been developed, and their performance on spike-in data sets has been evaluated previously. However, a comprehensive comparison of preprocessing algorithms on samples taken under research conditions has not been performed.
Methodology/principal findings: We used TaqMan RT-PCR arrays as a reference to evaluate the accuracy of expression values from Affymetrix microarrays in two experimental data sets: one comprising 84 genes in 36 colon biopsies, and the other comprising 75 genes in 29 cancer cell lines. We evaluated consistency using the Pearson correlation between measurements obtained on the two platforms. Also, we introduce the log-ratio discrepancy as a more relevant measure of discordance between gene expression platforms. Of nine preprocessing algorithms tested, PLIER+16 produced expression values that were most consistent with RT-PCR measurements, although the difference in performance between most of the algorithms was not statistically significant.
Conclusions/significance: Our results support the choice of PLIER+16 for the preprocessing of clinical Affymetrix microarray data. However, other algorithms performed similarly and are probably also good choices.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. The colon and cell line data sets are representative of clinical microarray data.
For several Affymetrix data sets, box-and-whiskers plots indicate the distribution of three bias metrics: a) RNA degradation slope, b) median perfect-match probe intensity, and c) fraction of probe sets called present. A narrower distribution indicates greater consistency in technical conditions. LatinSquare133 and LatinSquare95 are spike-in data sets produced by the microarray manufacturer ; Gyorffy_cells and Gyorffy_colon are the data sets analyzed in this paper , ; the other five are publicly-available clinical data sets –.
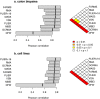
Figure 2. Pearson correlation coefficients between microarray and RT-PCR.
The distribution of Pearson correlation coefficients for each microarray preprocessing algorithm is indicated by a box plot, for a) the colon cancer data set (84 genes, 36 samples), and b) the cell line data set (75 genes, 29 samples). The box indicates the 25th to 75th percentile, and the heavier line indicates the median. Algorithms are displayed in decreasing order of the median, such that the more accurate algorithms are at the top. The colorgrams on the right-hand side indicate P values (Wilcoxon test) comparing each pair of algorithms.
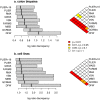
Figure 3. Log-ratio discrepancy between microarray and RT-PCR.
The distribution of the log-ratio discrepancy for each microarray preprocessing algorithm is indicated by a box plot, for a) the colon cancer data set, and b) the cell line data set. Algorithms are displayed in order of the median, such that the more accurate algorithms are at the top. The colorgrams on the right-hand side indicate P values (Wilcoxon test) comparing each pair of algorithms.
Similar articles
- Probe set filtering increases correlation between Affymetrix GeneChip and qRT-PCR expression measurements.
Mieczkowski J, Tyburczy ME, Dabrowski M, Pokarowski P. Mieczkowski J, et al. BMC Bioinformatics. 2010 Feb 24;11:104. doi: 10.1186/1471-2105-11-104. BMC Bioinformatics. 2010. PMID: 20181266 Free PMC article. - Comparing the use of Affymetrix to spotted oligonucleotide microarrays using two retinal pigment epithelium cell lines.
Rogojina AT, Orr WE, Song BK, Geisert EE Jr. Rogojina AT, et al. Mol Vis. 2003 Oct 6;9:482-96. Mol Vis. 2003. PMID: 14551534 Free PMC article. - Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study.
Arikawa E, Sun Y, Wang J, Zhou Q, Ning B, Dial SL, Guo L, Yang J. Arikawa E, et al. BMC Genomics. 2008 Jul 11;9:328. doi: 10.1186/1471-2164-9-328. BMC Genomics. 2008. PMID: 18620571 Free PMC article. - Validation of oligonucleotide microarray data using microfluidic low-density arrays: a new statistical method to normalize real-time RT-PCR data.
Abruzzo LV, Lee KY, Fuller A, Silverman A, Keating MJ, Medeiros LJ, Coombes KR. Abruzzo LV, et al. Biotechniques. 2005 May;38(5):785-92. doi: 10.2144/05385MT01. Biotechniques. 2005. PMID: 15945375 - Statistical challenges in preprocessing in microarray experiments in cancer.
Owzar K, Barry WT, Jung SH, Sohn I, George SL. Owzar K, et al. Clin Cancer Res. 2008 Oct 1;14(19):5959-66. doi: 10.1158/1078-0432.CCR-07-4532. Clin Cancer Res. 2008. PMID: 18829474 Free PMC article. Review.
Cited by
- Pathway-based evaluation in early onset colorectal cancer suggests focal adhesion and immunosuppression along with epithelial-mesenchymal transition.
Nam S, Park T. Nam S, et al. PLoS One. 2012;7(4):e31685. doi: 10.1371/journal.pone.0031685. Epub 2012 Apr 9. PLoS One. 2012. PMID: 22496728 Free PMC article. - Identifying disease-associated signaling pathways through a novel effector gene analysis.
Bao Z, Zhang B, Li L, Ge Q, Gu W, Bai Y. Bao Z, et al. PeerJ. 2020 Aug 14;8:e9695. doi: 10.7717/peerj.9695. eCollection 2020. PeerJ. 2020. PMID: 32864216 Free PMC article. - ToP: a trend-of-disease-progression procedure works well for identifying cancer genes from multi-state cohort gene expression data for human colorectal cancer.
Chung FH, Lee HH, Lee HC. Chung FH, et al. PLoS One. 2013 Jun 14;8(6):e65683. doi: 10.1371/journal.pone.0065683. Print 2013. PLoS One. 2013. PMID: 23799036 Free PMC article. - Inferring drug-disease associations from integration of chemical, genomic and phenotype data using network propagation.
Huang YF, Yeh HY, Soo VW. Huang YF, et al. BMC Med Genomics. 2013;6 Suppl 3(Suppl 3):S4. doi: 10.1186/1755-8794-6-S3-S4. Epub 2013 Nov 11. BMC Med Genomics. 2013. PMID: 24565337 Free PMC article. - The identification of gene expression profiles associated with progression of human diabetic neuropathy.
Hur J, Sullivan KA, Pande M, Hong Y, Sima AA, Jagadish HV, Kretzler M, Feldman EL. Hur J, et al. Brain. 2011 Nov;134(Pt 11):3222-35. doi: 10.1093/brain/awr228. Epub 2011 Sep 16. Brain. 2011. PMID: 21926103 Free PMC article.
References
- Affymetrix. 2008 Latin Square Data [ http://www.affymetrix.com/support/technical/sample_data/datasets.affx]
- Cope LM, Irizarry RA, Jaffee HA, Wu ZJ, Speed TP. A benchmark for affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–331. - PubMed
- Tong W, Lucas AB, Shippy R, Fan X, Fang H, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat Biotechnol. 2006;24:1132–1139. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases