Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans - PubMed (original) (raw)
. 2009 Jul 10;325(5937):197-201.
doi: 10.1126/science.1176225. Epub 2009 May 22.
C Todd Davis, Colin A Russell, Bo Shu, Stephen Lindstrom, Amanda Balish, Wendy M Sessions, Xiyan Xu, Eugene Skepner, Varough Deyde, Margaret Okomo-Adhiambo, Larisa Gubareva, John Barnes, Catherine B Smith, Shannon L Emery, Michael J Hillman, Pierre Rivailler, James Smagala, Miranda de Graaf, David F Burke, Ron A M Fouchier, Claudia Pappas, Celia M Alpuche-Aranda, Hugo López-Gatell, Hiram Olivera, Irma López, Christopher A Myers, Dennis Faix, Patrick J Blair, Cindy Yu, Kimberly M Keene, P David Dotson Jr, David Boxrud, Anthony R Sambol, Syed H Abid, Kirsten St George, Tammy Bannerman, Amanda L Moore, David J Stringer, Patricia Blevins, Gail J Demmler-Harrison, Michele Ginsberg, Paula Kriner, Steve Waterman, Sandra Smole, Hugo F Guevara, Edward A Belongia, Patricia A Clark, Sara T Beatrice, Ruben Donis, Jacqueline Katz, Lyn Finelli, Carolyn B Bridges, Michael Shaw, Daniel B Jernigan, Timothy M Uyeki, Derek J Smith, Alexander I Klimov, Nancy J Cox
Affiliations
- PMID: 19465683
- PMCID: PMC3250984
- DOI: 10.1126/science.1176225
Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans
Rebecca J Garten et al. Science. 2009.
Abstract
Since its identification in April 2009, an A(H1N1) virus containing a unique combination of gene segments from both North American and Eurasian swine lineages has continued to circulate in humans. The lack of similarity between the 2009 A(H1N1) virus and its nearest relatives indicates that its gene segments have been circulating undetected for an extended period. Its low genetic diversity suggests that the introduction into humans was a single event or multiple events of similar viruses. Molecular markers predictive of adaptation to humans are not currently present in 2009 A(H1N1) viruses, suggesting that previously unrecognized molecular determinants could be responsible for the transmission among humans. Antigenically the viruses are homogeneous and similar to North American swine A(H1N1) viruses but distinct from seasonal human A(H1N1).
Figures
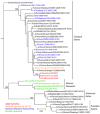
Fig. 1
Host and lineage origins for the gene segments of the 2009 A(H1N1) virus: PB2 polymerase basic 2, PB1 polymerase basic 1, PA polymerase acidic, HA hemagglutinin, NP nucleoprotein, NA neuraminidase, M matrix gene, NS nonstructural gene. Color of gene segment in circle indicates host. Determination of lineage explained in main text.
Fig. 2
A maximum likelihood phylogenetic tree for nucleotide sequences of the HA gene of influenza viruses selected to be representative of HA gene segments in relevant hosts. Phylogenetic trees of a larger number of representative HA gene segments, and of all H1 HA swine gene segments are shown in Figs. S1D and S2D respectively. Tree was inferred using PAUP* (version 4.0b10) (39), using GTR+I+Γ4 (the general time-reversible model with the proportion of invariant sites and the gamma distribution of among-site rate variation with four categories estimated from the empirical data) as determined by ModelTest (40). Global optimization of the tree topology was performed by tree bisection-reconnection branch swapping. The robustness of individual nodes of the tree was assessed using a bootstrap resampling analysis (1000 replicates, with topologies inferred using the neighbor-joining method under the GTR+I+Γ4 substitution model).
Fig. 3
Antigenic map of 71 early swine-origin 2009 A(H1N1) influenza viruses and 11 antisera. An antigenic map is a geometric representation of binding assay data, in this case the HI assay data in Tables S3 and S4. In such a map, the relative positions of strains (colored circles) and antisera (uncolored squares) are adjusted such that the distances between strains and antisera in the map represent the corresponding HI measurements with the least error. Distance in the map thus represents antigenic distance and the closer antigens are to each other in the map the more similar they are antigenically (37). The color of a circle in the map indicates whether the strain is a 2009 A(H1N1) influenza virus (blue) or an A(H1) swine influenza virus isolated between 1998 and 2007 from either a swine or a human infected with a swine influenza virus (green). The vertical and horizontal axes both represent antigenic distance, and because only the relative positions of antigens and antisera can be determined, the orientation of the map within these axes is free (thus an antigenic map can be rotated in the same way that a geographic map can be rotated). The spacing between grid lines is one unit of antigenic distance--corresponding to a 2-fold dilution of antiserum in the HI assay. Two units correspond to 4-fold dilution, three units to 8- fold dilution, etc. A difference higher than 4-fold in HI titer is usually considered to be sufficient to necessitate an update of the human seasonal influenza virus vaccine. Antigenic clusters of human seasonal influenza viruses typically have a radius of 2 antigenic units (4-fold in HI) (37). (see Fig. S3 for a zoomable PDF that includes the names of each strain and antiserum).
Similar articles
- Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013.
Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, Kelly M, Van Reeth K, Qiu Y, Simon G, Bonin E, Foni E, Chiapponi C, Larsen L, Hjulsager C, Markowska-Daniel I, Urbaniak K, Dürrwald R, Schlegel M, Huovilainen A, Davidson I, Dán Á, Loeffen W, Edwards S, Bublot M, Vila T, Maldonado J, Valls L; ESNIP3 Consortium; Brown IH, Pybus OG, Kellam P. Watson SJ, et al. J Virol. 2015 Oct;89(19):9920-31. doi: 10.1128/JVI.00840-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202246 Free PMC article. - The novel swine-origin H1N1 influenza A virus riddle: is it a domestic bird H1N1-derived virus?
Babakir-Mina M, Dimonte S, Ciccozzi M, Perno CF, Ciotti M. Babakir-Mina M, et al. New Microbiol. 2010 Jan;33(1):77-81. New Microbiol. 2010. PMID: 20402417 - Emergence and Evolution of Novel Reassortant Influenza A Viruses in Canines in Southern China.
Chen Y, Trovão NS, Wang G, Zhao W, He P, Zhou H, Mo Y, Wei Z, Ouyang K, Huang W, García-Sastre A, Nelson MI. Chen Y, et al. mBio. 2018 Jun 5;9(3):e00909-18. doi: 10.1128/mBio.00909-18. mBio. 2018. PMID: 29871917 Free PMC article. - [Swine influenza virus: evolution mechanism and epidemic characterization--a review].
Qi X, Lu C. Qi X, et al. Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese. - Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1).
Brockwell-Staats C, Webster RG, Webby RJ. Brockwell-Staats C, et al. Influenza Other Respir Viruses. 2009 Sep;3(5):207-13. doi: 10.1111/j.1750-2659.2009.00096.x. Influenza Other Respir Viruses. 2009. PMID: 19768134 Free PMC article. Review.
Cited by
- Synergistic Effect of S224P and N383D Substitutions in the PA of H5N1 Avian Influenza Virus Contributes to Mammalian Adaptation.
Song J, Xu J, Shi J, Li Y, Chen H. Song J, et al. Sci Rep. 2015 May 22;5:10510. doi: 10.1038/srep10510. Sci Rep. 2015. PMID: 26000865 Free PMC article. - Emergence of HA mutants during influenza virus pneumonia.
Manríquez ME, Makino A, Tanaka M, Abe Y, Yoshida H, Morioka I, Arakawa S, Takeshima Y, Iwata K, Takasaki J, Manabe T, Nakaya T, Nakamura S, Iglesias AL, Rossales RM, Mirabal EP, Ito T, Kitazawa T, Oka T, Yamashita M, Kudo K, Shinya K. Manríquez ME, et al. Int J Clin Exp Pathol. 2012;5(8):787-95. Epub 2012 Oct 1. Int J Clin Exp Pathol. 2012. PMID: 23071861 Free PMC article. - Secondary infection and clinical aspects after pandemic swine-origin influenza a (H1N1) admission in an Iranian critical care unite.
Hashemian SM, Tabarsi P, Nadji SA, Jamaati H, Mohajerani SA, Shamaee M, Chitsazan M, Radmand G, Maadani M, Mansouri SD. Hashemian SM, et al. Int J Crit Illn Inj Sci. 2014 Oct-Dec;4(4):309-13. doi: 10.4103/2229-5151.147536. Int J Crit Illn Inj Sci. 2014. PMID: 25625063 Free PMC article. - Pandemic Influenza A H1N1 (2009) Virus: Lessons from the Past and Implications for the Future.
Khanna M, Kumar B, Gupta A, Kumar P. Khanna M, et al. Indian J Virol. 2012 Jun;23(1):12-7. doi: 10.1007/s13337-012-0066-3. Epub 2012 Mar 25. Indian J Virol. 2012. PMID: 23729996 Free PMC article. - Age at Vaccination and Timing of Infection Do Not Alter Vaccine-Associated Enhanced Respiratory Disease in Influenza A Virus-Infected Pigs.
Souza CK, Rajão DS, Loving CL, Gauger PC, Pérez DR, Vincent AL. Souza CK, et al. Clin Vaccine Immunol. 2016 Jun 6;23(6):470-482. doi: 10.1128/CVI.00563-15. Print 2016 Jun. Clin Vaccine Immunol. 2016. PMID: 27030585 Free PMC article.
References
- Reid AH, Taubenberger JK. J Gen Virol. 2003;84:2285. - PubMed
- Sheerar MG, Easterday BC, Hinshaw VS. J Gen Virol. 1989;70(Pt 12):3297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous