Heterologous cross-seeding mimics cross-species prion conversion in a yeast model - PubMed (original) (raw)
Heterologous cross-seeding mimics cross-species prion conversion in a yeast model
Namitha Vishveshwara et al. BMC Biol. 2009.
Abstract
Background: Prions are self-perpetuating, infectious, aggregated proteins that are associated with several neurodegenerative diseases in mammals and heritable traits in yeast. Sup35p, the protein determinant of the yeast prion [PSI+], has a conserved C terminal domain that performs the Sup35p function and a prion domain that is highly divergent. Prions formed by chimeras of the prion domain of various species fused to the C domain of Saccharomyces cerevisiae exhibit a 'species barrier', a phenomenon first observed in mammals, and often fail to transmit the prion state to chimeras with prion domains of other species.
Results: We focus on the chimera containing the prion domain of Pichia methanolica and examine how tight the 'species barrier' is between the chimera and S. cerevisiae. Although either of two Q/N-rich prions, [PSI+] or [PIN+], enhances the formation of the chimeric prion, [CHI+PM], neither a non-Q/N-rich prion nor a non-prion Q-rich aggregate promotes the formation of [CHI+PM]. [CHI+PM] has many features characteristic of yeast prions: aggregation, cytoplasmic transmission and a two-level protein structure. [CHI+PM] formed in the presence of [PSI+] can propagate independently of [PSI+] and forms at least two different variants of the prion, suggesting the generation and not transmission of new prion seeds.
Conclusion: Although the sequence similarity between the S. cerevisiae Q/N-rich prion determinants and the P. methanolica prion domain is low, we find that the chimera containing the prion domain of P. methanolica can occasionally be cross-seeded by [PSI+] to mimic crossing the species barrier, to form the [CHI+PM] prion. Our data suggests that crossing the barrier occurs by a de novo formation of the foreign chimeric prion. Thus, the species barrier appears to be crossed by a heterologous seeding mechanism, wherein the infected prion protein uses the pre-existing seed as an inefficient template.
Figures
Figure 1
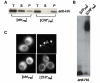
NMPM-CSC but not NMCA-CSC is inactivated in the presence of [PSI+]. A. NMPM-CSC and NMCA-CSC provide translation termination function. [PSI+] or sup35 cells ectopically expressing prion domains of either Pichia methanolica (NMPM) or Candida albicans (NMCA) fused to the C domain of Saccharomyces cerevisiae (NMSC) expressed on a URA3 plasmid on media containing low adenine-Ura and -Ade-Ura. -Ura is used to maintain the plasmid. B. NMPM-CSC is inactivated in [PSI+] cells and not in a sup35 mutant. Around 107 [PSI+] cells expressing either NMPM-CSC or NMCA-CSC, or sup35 mutant yeast expressing NMPM-CSC were plated on -Ade-Ura media.
Figure 2
Ade+ colonies have prion properties. A. NMPM-CSC is mainly in the pellet fraction in Ade+ colonies. [CHI+PM] and [_chi_-PM] lysates subjected to high-speed centrifugation and probed for the chimeric protein using an anti-HA tag antibody. T: total; S: supernatant; P: pellet. B. [CHI+PM] colonies have SDS-stable sub-particles. [CHI+PM] and [_chi_-PM] lysates analyzed on an agarose gel and probed for the chimeric protein (anti-HA tag). C. NMPM-GFP forms puncta in [CHI+PM] colonies. NMPM-GFP under a copper-inducible promoter overexpressed in [CHI+PM] or [_chi_-PM] cells for 4 hours and examined under a fluorescence microscope. The two kinds of NMPM-GFP puncta are sometimes observable in the same [CHI+PM] culture and are not representative of different [CHI+PM] colonies.
Figure 3
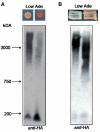
NMPM-CSC is not incorporated into [PSI+] sub-particles. [_chi_-PM][PSI+] and [CHI+PM][PSI+] lysates resolved on an agarose gel and probed for the chimeric protein (anti-HA) and reprobed for endogenous Sup35p (_anti-NM_SC).
Figure 4
More than one variant of [CHI+PM] forms in the presence of [PSI+]. A. Some [CHI+PM] variants have differing amounts of SDS-stable sub-particles. Lysates from [CHI+PM] colonies differing in color on low adenine media were resolved on an agarose gel and probed with an anti-HA tag antibody that detects NMPM-CSC. Stained chicken pectoralis muscle extract provided molecular weight markers. The difference in amount of SDS-stable sub-particles was reproducible for this and another pair (not shown) isolated on the basis of color difference. B. Some [CHI+PM] variants have different sized SDS-stable sub-particles. Colonies formed on -Ade-Ura could be distinguished phenotypically on low adenine media by differences in color. Lysates from these variants were resolved on an agarose gel and probed with anti-HA tag antibody, which detects NMPM-CSC. The difference in size of SDS-stable sub-particles was reproducible for this and another pair (not shown) isolated on the basis of color difference.
Figure 5
[PIN+] enhances [CHI+PM] formation. A. NMPM-CSC provides translation termination function to a sup35 mutant containing the medium [PIN+] variant. sup35 medium [PIN+] or [_pin_-] yeast ectopically expressing NMPM-CSC on media containing low or no adenine without uracil (to maintain the plasmid). Cells with the medium [PIN+] variant are shown here. NMPM-CSC is also functional in sup35 mutant yeast with the low [PIN+] variant. B. NMPM-CSC is inactivated in sup35 mutant cells containing the medium or low [PIN+] variant and not in a [_pin_-] sup35 mutant Around 107 sup35 mutant cells containing either medium [PIN+] or [_pin_-] expressing NMPM-CSC were plated onto -Ade-Ura. The frequency of Ade+ colonies was estimated in sup35 yeast with medium and low [PIN+] variants. The Ade+ frequency in sup35 yeast with high and very high [PIN+] variants was not determined as suppression in these strains is enhanced compared with sup35 [_pin_-] yeast.
Similar articles
- Prion species barrier between the closely related yeast proteins is detected despite coaggregation.
Chen B, Newnam GP, Chernoff YO. Chen B, et al. Proc Natl Acad Sci U S A. 2007 Feb 20;104(8):2791-6. doi: 10.1073/pnas.0611158104. Epub 2007 Feb 12. Proc Natl Acad Sci U S A. 2007. PMID: 17296932 Free PMC article. - Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein.
Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Chernoff YO, et al. Mol Microbiol. 2000 Feb;35(4):865-76. doi: 10.1046/j.1365-2958.2000.01761.x. Mol Microbiol. 2000. PMID: 10692163 - Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro.
Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Derkatch IL, et al. Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):12934-9. doi: 10.1073/pnas.0404968101. Epub 2004 Aug 23. Proc Natl Acad Sci U S A. 2004. PMID: 15326312 Free PMC article. - Cellular factors important for the de novo formation of yeast prions.
Tuite M, Stojanovski K, Ness F, Merritt G, Koloteva-Levine N. Tuite M, et al. Biochem Soc Trans. 2008 Oct;36(Pt 5):1083-7. doi: 10.1042/BST0361083. Biochem Soc Trans. 2008. PMID: 18793193 Review. - Interactions between non-identical prion proteins.
Gonzalez Nelson AC, Ross ED. Gonzalez Nelson AC, et al. Semin Cell Dev Biol. 2011 Jul;22(5):437-43. doi: 10.1016/j.semcdb.2011.02.022. Epub 2011 Feb 24. Semin Cell Dev Biol. 2011. PMID: 21354317 Review.
Cited by
- Prions in yeast.
Liebman SW, Chernoff YO. Liebman SW, et al. Genetics. 2012 Aug;191(4):1041-72. doi: 10.1534/genetics.111.137760. Genetics. 2012. PMID: 22879407 Free PMC article. Review. - The [PSI+] prion exists as a dynamic cloud of variants.
Bateman DA, Wickner RB. Bateman DA, et al. PLoS Genet. 2013;9(1):e1003257. doi: 10.1371/journal.pgen.1003257. Epub 2013 Jan 31. PLoS Genet. 2013. PMID: 23382698 Free PMC article. - Site-specific structural analysis of a yeast prion strain with species-specific seeding activity.
Marcelino-Cruz AM, Bhattacharya M, Anselmo AC, Tessier PM. Marcelino-Cruz AM, et al. Prion. 2011 Jul-Sep;5(3):208-14. doi: 10.4161/pri.5.3.16694. Epub 2011 Jul 1. Prion. 2011. PMID: 22048721 Free PMC article. - β-amyloid oligomers and prion protein: Fatal attraction?
Forloni G, Balducci C. Forloni G, et al. Prion. 2011 Jan-Mar;5(1):10-5. doi: 10.4161/pri.5.1.14367. Epub 2011 Jan 1. Prion. 2011. PMID: 21150333 Free PMC article. - Amyloids and yeast prion biology.
Wickner RB, Edskes HK, Bateman DA, Kelly AC, Gorkovskiy A, Dayani Y, Zhou A. Wickner RB, et al. Biochemistry. 2013 Mar 5;52(9):1514-27. doi: 10.1021/bi301686a. Epub 2013 Feb 12. Biochemistry. 2013. PMID: 23379365 Free PMC article.
References
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. - PubMed
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. - PubMed
- Collinge J. Variant Creutzfeldt-Jakob disease. Lancet. 1999;354:317–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases