Studies on the DIDS-binding site of monocarboxylate transporter 1 suggest a homology model of the open conformation and a plausible translocation cycle - PubMed (original) (raw)
Studies on the DIDS-binding site of monocarboxylate transporter 1 suggest a homology model of the open conformation and a plausible translocation cycle
Marieangela C Wilson et al. J Biol Chem. 2009.
Abstract
Site-directed mutagenesis of MCT1 was performed on exofacial lysines Lys(38), Lys(45), Lys(282), and Lys(413). K38Q-MCT1 and K38R-MCT1 were inactive when expressed at the plasma membrane of Xenopus laevis oocytes, whereas K45R/K282R/K413R-MCT1 and K45Q/K282Q/K413Q-MCT1 were active. The former exhibited normal reversible and irreversible inhibition by DIDS, whereas the latter showed less reversible and no irreversible inhibition. K45Q/K413Q-MCT1 retained some irreversible inhibition, whereas K45Q/K282Q-MCT1 and K282Q/K413Q-MCT1 did not. These data suggest that the two DIDS SO(3)(-) groups interact with positively charged Lys(282) together with Lys(45) and/or Lys(413). This positions one DIDS isothiocyanate group close to Lys(38), leading to its covalent modification and irreversible inhibition. Additional mutagenesis revealed that DIDS cross-links MCT1 to its ancillary protein embigin using either Lys(38) or Lys(290) of MCT1 and Lys(160) or Lys(164) of embigin. We have modeled a possible structure for the outward facing (open) conformation of MCT1 by employing modest rotations of the C-terminal domain of the inner facing conformation modeled previously. The resulting model structure has a DIDS-binding site consistent with experimental data and locates Lys(38) in a hydrophobic environment at the bottom of a substrate-binding channel. Our model suggests a translocation cycle in which Lys(38) accepts a proton before binding lactate. Both the lactate and proton are then passed through the channel via Asp(302-) and Asp(306+), an ion pair already identified as important for transport and located adjacent to Phe(360), which controls channel selectivity. The cross-linking data have also been used to model a structure of MCT1 bound to embigin that is consistent with published data.
Figures
FIGURE 1.
Lys38 is essential for MCT1 activity. Xenopus oocytes were injected with water or cRNA for wild type MCT1 or the mutant shown, and after expression for 3 days the uptake of
l
-[14C]lactate was determined as describe under “Experimental Procedures.” Where indicated the oocytes were treated with 0.2 m
m
DIDS under conditions designed to reveal reversible or irreversible inhibition. The conditions were chosen that gave incomplete inhibition so that either increases or decreases in inhibitor affinity caused by subsequent mutations could be readily detected. The data are shown as the means ± S.E. of 30–180 separate oocytes. The insets show a Western blot of crude membrane fractions (equal protein loading) and sections of oocytes subject to immunofluorescence microscopy to confirm that the lack of lactate transport by the K38Q (inset B) or K38R (inset C) mutants is not due to reduced MCT1 expression compared with wild type MCT1 (inset A). Inset D is water-injected. Scale bar, 20 μ
m
.
FIGURE 2.
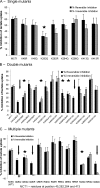
The effect of lysine mutations of MCT1 on the reversible and irreversible inhibition of lactate transport by DIDS. Lactate transport by oocytes expressing MCT1 with single (A) double (B) or multiple (C) mutations of lysines to glutamines or arginines was measured in the absence or presence of DIDS under conditions designed to reveal reversible (solid bars) or irreversible inhibition (shaded bars) as described under “Experimental Procedures.” The data are expressed as percentages of inhibition of transport by DIDS and as the means ± S.E. of at least 12 oocytes. All of these mutant forms of MCT1 were active and supported rates of lactate transport within 45–120% of wild type (WT) MCT1 as reported in
supplemental Table S2
. Mutations showing a significant reduction in reversible and irreversible inhibition (p < 0.05 by Student's t test) are indicated with a star, whereas those in which irreversible inhibition is abolished are indicated with an arrow.
FIGURE 3.
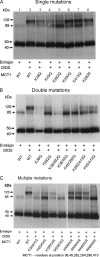
The effect of lysine mutations of MCT1 on the DIDS-mediated cross-linking of MCT1 to embigin. COS cells were transfected with the indicated pCI_neo_ MCT1 and embigin constructs and harvested after 48 h before incubating with or without 0.2 m
m
DIDS at 37 °C for 1 h. The cells were then washed and lysed to produce crude lysates, which were subject to SDS-PAGE and Western blotting with anti-MCT1 antibody as described under “Experimental Procedures.” The bands at ∼45 and 85 kDa represent monomeric and dimeric MCT1, whereas the band at ∼120 kDa represents the cross-linked product of MCT1 with embigin. The data shown are representative of two or three experiments that gave similar results. WT, wild type.
FIGURE 4.
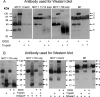
The effects of trypsin digestion of DIDS-treated rat erythrocyte ghosts on the cross-linked product of MCT1 with embigin. In panel A, rat erythrocytes were incubated in the absence or presence of 100 μ
m
DIDS for 1 h at 37 °C prior to preparation of ghosts and, where required, treatment with trypsin as described under “Experimental Procedures.” The proteins were subjected to SDS-PAGE and Western blotting using the antibodies against the C terminus or 7/8 loop of MCT1 or an antibody raised against the cross-linked product of MCT1 with embigin (16). In B data are shown for samples that have been treated with _N_-glycanase F (DGaseF) as described under “Experimental Procedures.” The data shown are representative of two or three experiments that gave similar results.
FIGURE 5.
Schematic representation of trypsin cleavage and antibody binding sites on MCT1 and the probable locus of cross-linking between MCT1 and embigin.
FIGURE 6.
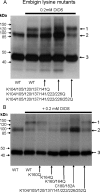
Lysine residues 160 and 164 of embigin are involved in DIDS-mediated cross-linking to MCT1. COS cells were transfected as detailed under “Experimental Procedures” with the following pCIneo vector constructs. A, first and second lanes, MCT1 + embigin; third lane, MCT1 + K104/105/120/137/141Q embigin; fourth lane, MCT1 + K104/105/120/137/141/222/226Q embigin; fifth lane, MCT1 + K104/105/120/137/141/222/226/252Q embigin. Arrows 1–3 indicate the positions of MCT1-embigin cross-linked product, MCT1 dimer, and MCT1 monomer, respectively. B, MCT1 plus the embigin mutant indicated. COS cells were harvested 48 h after transfection and treated ± 0.2 m
m
DIDS, washed, and lysed. SDS-PAGE and Western blots were performed on the crude lysates using anti-MCT1 antibody. WT, wild type.
FIGURE 7.
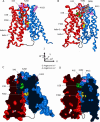
MCT1 models. A and C, closed; B and D, open. The N-terminal domain is colored red, and the C-terminal domain colored blue; Lys38 is green, and Lys45, Lys282, and Lys413 are pink, and Phe360 is yellow, shown in space-filling representation. The 30-residue loop connecting the N- and C-terminal domains is indicated by the sketched black line (not modeled). A and B are ribbon diagrams of the closed and open models, respectively. A space-filling model of DIDS in gray shows the proposed binding site in the open conformation (B). C and D show the same structures as A and B but are cross-sections of the models rendered with a solvent-accessible surface. Hence C shows how the TM channel is closed on the extracellular (upper) side, whereas D show the open TM channel, gated by Lys38 with the DIDS-binding site on the extracellular side of this gate. The position of the specificity filter (Asp302, Arg306, and Phe360) is indicated by the position of Phe360 (Asp302 and Arg306 are not shown for clarity, but line the channel next to Phe360). The axis system used for the C-terminal domain rotations and translations to generate the open model is shown in the center of the figure.
FIGURE 8.
Cartoon illustrating the proposed mechanism of lactic acid transport by MCT1. Lactic acid protonates Lys38 (K) causing the channel to open. Lactate then moves into the open extracellular side of the pore and forms an ion pair with Lys38. In the next step the proton on Lys38 is transferred to aspartate 302 (_D_-) neutralizing the aspartate side chain (DH). This is followed by migration of lactate through the pore where it forms an ion pair with R306 (R+). The size of the adjacent side chain of residue 360 (F) governs the ability of this site to bind the α-hydroxy acid. Once Lys38 is deprotonated, and lactate is occupying the specificity filter, the transporter relaxes back toward the closed state and releases lactic acid into the intracellular space.
FIGURE 9.
The binding of DIDS to the open conformation of MCT1. The figure shows a side (A) and top (B) view of the extracellular part of the open MCT1 model. The color scheme is the same as that in Fig. 7, except that DIDS is colored according to atom type with the carbon atoms colored gray. The figure shows how one isothiocyanate group of DIDS is in close contact with Lys38, whereas the two sulfonate groups are bound to Lys45, Lys282, and Lys413. The inset in B shows the numbering of the TM helices in this view.
FIGURE 10.
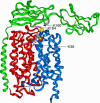
Structural model of MCT1 associated with embigin illustrating the site of DIDS cross-linking. Details of the modeling strategy are given in the text. The proteins are represented as ribbons (red, N-MCT1; blue, C-MCT1; green, embigin). The lysine residues involved in cross-linking (Lys38 of MCT1, and Lys164 and Lys160 of embigin) are indicated as magenta sticks, whereas the cysteine disulfide bridges of the two immunoglobulin domains of embigin are shown as yellow sticks. DIDS is shown as a space-filling representation, with standard atom coloring.
Similar articles
- Identification of key binding site residues of MCT1 for AR-C155858 reveals the molecular basis of its isoform selectivity.
Nancolas B, Sessions RB, Halestrap AP. Nancolas B, et al. Biochem J. 2015 Feb 15;466(1):177-88. doi: 10.1042/BJ20141223. Biochem J. 2015. PMID: 25437897 Free PMC article. - The inhibition of monocarboxylate transporter 2 (MCT2) by AR-C155858 is modulated by the associated ancillary protein.
Ovens MJ, Manoharan C, Wilson MC, Murray CM, Halestrap AP. Ovens MJ, et al. Biochem J. 2010 Oct 15;431(2):217-25. doi: 10.1042/BJ20100890. Biochem J. 2010. PMID: 20695846 Free PMC article. - The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity.
Manoharan C, Wilson MC, Sessions RB, Halestrap AP. Manoharan C, et al. Mol Membr Biol. 2006 Nov-Dec;23(6):486-98. doi: 10.1080/09687860600841967. Mol Membr Biol. 2006. PMID: 17127621 Free PMC article. - Monocarboxylate Transporter 1 in Brain Diseases and Cancers.
Sun Y, Sun J, He Z, Wang G, Wang Y, Zhao D, Wang Z, Luo C, Tian C, Jiang Q. Sun Y, et al. Curr Drug Metab. 2019;20(11):855-866. doi: 10.2174/1389200220666191021103018. Curr Drug Metab. 2019. PMID: 31631816 Review. - The SLC16 gene family - structure, role and regulation in health and disease.
Halestrap AP. Halestrap AP. Mol Aspects Med. 2013 Apr-Jun;34(2-3):337-49. doi: 10.1016/j.mam.2012.05.003. Mol Aspects Med. 2013. PMID: 23506875 Review.
Cited by
- Monocarboxylate Transporters: Therapeutic Targets and Prognostic Factors in Disease.
Jones RS, Morris ME. Jones RS, et al. Clin Pharmacol Ther. 2016 Nov;100(5):454-463. doi: 10.1002/cpt.418. Epub 2016 Aug 22. Clin Pharmacol Ther. 2016. PMID: 27351344 Free PMC article. Review. - Direct cell-to-cell transfer in stressed tumor microenvironment aggravates tumorigenic or metastatic potential in pancreatic cancer.
Jang G, Oh J, Jun E, Lee J, Kwon JY, Kim J, Lee SH, Kim SC, Cho SY, Lee C. Jang G, et al. NPJ Genom Med. 2022 Oct 27;7(1):63. doi: 10.1038/s41525-022-00333-w. NPJ Genom Med. 2022. PMID: 36302783 Free PMC article. - The structural insight into the functional modulation of human anion exchanger 3.
Jian L, Zhang Q, Yao D, Wang Q, Chen M, Xia Y, Li S, Shen Y, Cao M, Qin A, Li L, Cao Y. Jian L, et al. Nat Commun. 2024 Jul 20;15(1):6134. doi: 10.1038/s41467-024-50572-x. Nat Commun. 2024. PMID: 39033175 Free PMC article. - Juvenile cataract-associated mutation of solute carrier SLC16A12 impairs trafficking of the protein to the plasma membrane.
Castorino JJ, Gallagher-Colombo SM, Levin AV, Fitzgerald PG, Polishook J, Kloeckener-Gruissem B, Ostertag E, Philp NJ. Castorino JJ, et al. Invest Ophthalmol Vis Sci. 2011 Aug 29;52(9):6774-84. doi: 10.1167/iovs.10-6579. Invest Ophthalmol Vis Sci. 2011. PMID: 21778275 Free PMC article. - S-Nitrosation of monocarboxylate transporter 1: inhibition of pyruvate-fueled respiration and proliferation of breast cancer cells.
Diers AR, Broniowska KA, Chang CF, Hill RB, Hogg N. Diers AR, et al. Free Radic Biol Med. 2014 Apr;69:229-38. doi: 10.1016/j.freeradbiomed.2014.01.031. Epub 2014 Jan 30. Free Radic Biol Med. 2014. PMID: 24486553 Free PMC article.
References
- Halestrap A. P., Meredith D. (2004) Pflugers Arch. 447, 619–628 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases