Human caspases: activation, specificity, and regulation - PubMed (original) (raw)
Review
. 2009 Aug 14;284(33):21777-21781.
doi: 10.1074/jbc.R800084200. Epub 2009 May 26.
Affiliations
- PMID: 19473994
- PMCID: PMC2755903
- DOI: 10.1074/jbc.R800084200
Review
Human caspases: activation, specificity, and regulation
Cristina Pop et al. J Biol Chem. 2009.
Abstract
Caspases are intracellular proteases that propagate programmed cell death, proliferation, and inflammation. Activation of caspases occurs by a conserved mechanism subject to strict cellular regulation. Once activated by a specific stimulus, caspases execute limited proteolysis of downstream substrates to trigger a cascade of events that culminates in the desired biological response. Much has been learned of the mechanisms that govern the activation and regulation of caspases, and this minireview provides an update of these areas. We also delineate substantial gaps in knowledge of caspase function, which can be approached by techniques and experimental paradigms that are currently undergoing development.
Figures
FIGURE 1.
A, caspase organization. A prodomain precedes the catalytic domain, composed of two covalently linked subunits. Sites for (auto)proteolysis at Asp residues are indicated. B, activation mechanisms. Initiators are monomers that activate by prodomain-mediated dimerization. Executioners are dimers that activate by cleavage of intersubunit linkers. Following activation, additional proteolytic events mature the caspases to more stable forms, prone to regulation.
FIGURE 2.
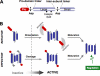
A, activation pathways and substrates. An oligomeric protein platform activates an apical caspase (casp), which then cleaves specific caspases. The apoptotic apical caspases require an intermediary step through the direct activation of downstream caspases, creating a two-step pathway that amplifies the apoptotic signal and allows for additional regulation points. A few caspase substrates are shown, but many are yet to be discovered. Caspase inhibitors (shown in boxes) regulate the activation pathways. IL, interleukin; ICAD, inhibitor of caspase-activated DNase; PARP, poly(ADP-ribose) polymerase; Rb, retinoblastoma protein; _PKC_δ, protein kinase Cδ. B, substrate specificity of caspases. The figure plots amino acid preferences in the P4–P1′ positions of caspases. The total height of each position is proportional to sequence conservation, whereas the height of residues in each position plots the relative frequency of each amino acid at the position (see the WebLogo web site). The Casp-3 and Casp-8 panels summarize peptide libraries exploring the P4–P1′ substrate primary specificity determinants and reveal little overlap between caspase-3 and -8 (taken from data in Refs. and 35). The Apoptotic cells panel shows caspase substrates from natural proteins cleaved in apoptotic cells, revealing an almost total dropout in specificity of the P4 site, implying that selectivity for natural substrates may fail to obey the simplistic rules established with synthetic peptide library substrates (taken from data in Ref. 18).
Similar articles
- Caspases in cell survival, proliferation and differentiation.
Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Lamkanfi M, et al. Cell Death Differ. 2007 Jan;14(1):44-55. doi: 10.1038/sj.cdd.4402047. Epub 2006 Oct 20. Cell Death Differ. 2007. PMID: 17053807 Review. - Caspases as regulators of apoptosis and other cell functions.
Philchenkov AA. Philchenkov AA. Biochemistry (Mosc). 2003 Apr;68(4):365-76. doi: 10.1023/a:1023635510363. Biochemistry (Mosc). 2003. PMID: 12765517 Review. - Caspases and their substrates.
Julien O, Wells JA. Julien O, et al. Cell Death Differ. 2017 Aug;24(8):1380-1389. doi: 10.1038/cdd.2017.44. Epub 2017 May 12. Cell Death Differ. 2017. PMID: 28498362 Free PMC article. Review. - Caspases and apoptosis.
Salvesen GS. Salvesen GS. Essays Biochem. 2002;38:9-19. doi: 10.1042/bse0380009. Essays Biochem. 2002. PMID: 12463158 Review. - Caspases at the crossroads of immune-cell life and death.
Siegel RM. Siegel RM. Nat Rev Immunol. 2006 Apr;6(4):308-17. doi: 10.1038/nri1809. Nat Rev Immunol. 2006. PMID: 16557262 Review.
Cited by
- Genuine selective caspase-2 inhibition with new irreversible small peptidomimetics.
Bosc E, Anastasie J, Soualmia F, Coric P, Kim JY, Wang LQ, Lacin G, Zhao K, Patel R, Duplus E, Tixador P, Sproul AA, Brugg B, Reboud-Ravaux M, Troy CM, Shelanski ML, Bouaziz S, Karin M, El Amri C, Jacotot ED. Bosc E, et al. Cell Death Dis. 2022 Nov 15;13(11):959. doi: 10.1038/s41419-022-05396-2. Cell Death Dis. 2022. PMID: 36379916 Free PMC article. - Mechanisms of cell death in heart disease.
Konstantinidis K, Whelan RS, Kitsis RN. Konstantinidis K, et al. Arterioscler Thromb Vasc Biol. 2012 Jul;32(7):1552-62. doi: 10.1161/ATVBAHA.111.224915. Epub 2012 May 17. Arterioscler Thromb Vasc Biol. 2012. PMID: 22596221 Free PMC article. Review. - Interactions between mitochondria and the transcription factor myocyte enhancer factor 2 (MEF2) regulate neuronal structural and functional plasticity and metaplasticity.
Brusco J, Haas K. Brusco J, et al. J Physiol. 2015 Aug 15;593(16):3471-81. doi: 10.1113/jphysiol.2014.282459. Epub 2015 Jan 29. J Physiol. 2015. PMID: 25581818 Free PMC article. Review. - Maspin is a deoxycholate-inducible, anti-apoptotic stress-response protein differentially expressed during colon carcinogenesis.
Payne CM, Holubec H, Crowley-Skillicorn C, Nguyen H, Bernstein H, Wilcox G, Bernstein C. Payne CM, et al. Clin Exp Gastroenterol. 2011;4:239-53. doi: 10.2147/CEG.S24093. Epub 2011 Oct 3. Clin Exp Gastroenterol. 2011. PMID: 22162927 Free PMC article. - The caspase 6 derived N-terminal fragment of DJ-1 promotes apoptosis via increased ROS production.
Robert G, Puissant A, Dufies M, Marchetti S, Jacquel A, Cluzeau T, Colosetti P, Belhacene N, Kahle P, Da Costa CA, Luciano F, Checler F, Auberger P. Robert G, et al. Cell Death Differ. 2012 Nov;19(11):1769-78. doi: 10.1038/cdd.2012.55. Epub 2012 May 4. Cell Death Differ. 2012. PMID: 22555455 Free PMC article.
References
- Yuan J., Shaham S., Ledoux S., Ellis H. M., Horvitz H. R. (1993) Cell 75, 641–652 - PubMed
- Lamkanfi M., Festjens N., Declercq W., Vanden Berghe T., Vandenabeele P. (2007) Cell Death Differ. 14, 44–55 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases