Functional and evolutionary insights into human brain development through global transcriptome analysis - PubMed (original) (raw)
Functional and evolutionary insights into human brain development through global transcriptome analysis
Matthew B Johnson et al. Neuron. 2009.
Abstract
Our understanding of the evolution, formation, and pathological disruption of human brain circuits is impeded by a lack of comprehensive data on the developing brain transcriptome. A whole-genome, exon-level expression analysis of 13 regions from left and right sides of the mid-fetal human brain revealed that 76% of genes are expressed, and 44% of these are differentially regulated. These data reveal a large number of specific gene expression and alternative splicing patterns, as well as coexpression networks, associated with distinct regions and neurodevelopmental processes. Of particular relevance to cognitive specializations, we have characterized the transcriptional landscapes of prefrontal cortex and perisylvian speech and language areas, which exhibit a population-level global expression symmetry. We show that differentially expressed genes are more frequently associated with human-specific evolution of putative cis-regulatory elements. These data provide a wealth of biological insights into the complex transcriptional and molecular underpinnings of human brain development and evolution.
Figures
Figure 1. Regional and Neocortical Areal Gene Expression Profiling in the Mid-Fetal Human Brain
(A) Human late mid-fetal brain illustrating locations of tissue samples microdissected from nine areas of neocortex (NCTX; including four areas of the prefrontal cortex [PFC]), hippocampus (HIP), striatum (STR), mediodorsal thalamus (THM), and cerebellum (CBL) from both sides of four mid-fetal brains. Dashed lines (i, ii) indicate levels of acetylcholine esterase-reacted coronal tissue sections. (B) Comparison among NCTX, HIP, STR, THM, and CBL detected 76% of core genes expressed above background in at least one brain region. At a FDR of 10−5, 33% of these were DEX and 28% DAS between regions. (C) Intra-NCTX analysis yielded fewer DEX and DAS genes, even at a relaxed threshold (FDR = 0.01).
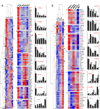
Figure 2. Unsupervised Hierarchical Clustering and qRT-PCR Validation of Selected Genes Differentially Expressed Between Brain Regions
Genes with FDR<10−5 and greater than twofold maximum expression difference were clustered using uncentered correlation and average linkage. Selected highly correlated (r>0.75) clusters of genes with the most restricted expression patterns, indicated in the heatmap at left, are shown in detail at right, with representative validated genes labeled in red. Red is higher expression, blue is lower expression. Complete gene lists for these clusters are given in Table S6. Bar graphs show qRT-PCR (black bars) next to Exon Array data (grey bars) for representative validated genes. Bar graphs represent fold-change relative to the average of all samples for qRT-PCR (black bars; mean±SEM) and median normalized log expression level for Exon Array (grey bars).
Figure 3. Unsupervised Hierarchical Clustering and qRT-PCR Validation of Selected Genes Differentially Expressed Between NCTX Areas
(A) With PFC areas grouped, correlated clusters (r>0.8) of gene enrichment included: (1 and 2) PFC; (3) temporal (TAU+TAS) +OCC; (4) temporal lobe (TAS+TAU); (5) PFC+TAS; and (6) OCC. (B) We separately analyzed the four PFC areas plus MS, and identified genes enriched within specific frontal lobe areas. Selected clusters include (1) pan-PFC, but not MS; (2) OPFC; (3) orbital and lateral PFC; (4) MPFC; (5) MPFC+MS; (6) VLPFC+MS. Red is higher expression, blue is lower expression. Bar graphs show qRT-PCR confirmation (black bars; mean±SEM) next to array data (grey bars) (axes as in Figure 2) with a median ANOVA p<0.0004 and correlation to array data r=0.9. Complete lists of genes in these clusters are given in Table S7–Table S8.
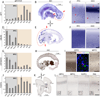
Figure 4. Confirmation and Cellular Mapping of Selected Neocortical Areal Expression Differences
Confirmation of array results by qRT-PCR, and the detection of areal- and cell type-restricted expression patterns by ISH and IHC. (A–C) NPY enrichment in non-frontal areas is confirmed by qRT-PCR, with highest expression in temporal (TAS, TAU) and occipital lobes (OCC). (B) ISH on a whole sagittal section of 24 wg human brain confirms NPY enrichment in OCC. NPY enrichment in temporal cortex is not visible in this very medial tissue section. (C) Higher magnification reveals specific enrichment in the middle of the occipital cortical plate (CP), and high expression in scattered cells throughout the subplate (SP) (red arrowheads). (D–F) CBLN2 enrichment in OPFC and lateral PFC is confirmed by qRT-PCR (D) and ISH on a whole sagittal section of 24 wg brain (E). (F) Higher magnification reveals that CBLN2 is enriched throughout the prefrontal CP and SP, but absent from the marginal zone (MZ). (G–I) CNTNAP2 is selectively enriched in OPFC and lateral PFC areas. (H–I) IHC reveals specific diffuse enrichment in orbitofrontal SP and high expression in scattered MZ cells. Triple-immunofluorescent staining (I, middle panel) reveals colocalization of CNTNAP2 with astrocytic marker GFAP but not neuronal marker NeuN, suggesting differential expression of CNTNAP2 in SP astrocytes. (J–L) FOXP2 is differentially expressed within the frontal cortex, and enriched in perisylvian cortex. IHC in coronal 22 wg brain sections (K–L) suggests that these differences are accounted for by a combination of higher cellular expression levels in the CP, particularly in OPFC (lower insets), and greater numbers of FOXP2-immunopositive SP cells, especially in VLPFC and PAS. Interestingly, strongly FOXP2-positive cells were present in the MZ in VLPFC and OPFC, but were completely absent from the MZ in other areas (upper insets). Bar graphs are mean±SEM. Scale bars in C, F, I, L: 500 µm.
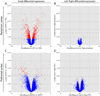
Figure 5. Population-level Global Left-Right Symmetry of Gene Expression in the Late Mid-Fetal Prefrontal and Perisylvian Neocortex
Volcano plots depicting the range of fold-differences and uncorrected p-values in representative perisylvian (A–B) and prefrontal (C–D) areas of NCTX. A large number of genes show significant differences in expression between NCTX areas (A and C). In contrast, no genes showed significant differences between left and right hemispheres within areas (B and D). Dashed lines indicate a corrected p-value of 0.01 (step-up FDR).
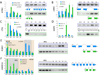
Figure 6. Validation of Selected Region-Specific Alternative Splicing Patterns
Validation of differential AS across brain regions (A–D) or neocortical areas (E–F) by exon- or isoform-specific qRT-PCR. (A) Confirmation of a specific reduction of truncated NTRK2b in NCTX, whereas full-length NTRK2a is more evenly expressed throughout the brain. (B) The LIMK2b splice variant, which lacks the N-terminal LIM protein-binding domain, is predominantly enriched in THM and CBL. (C–D) CPVL and SKAP2 are each predicted to encode a previously unknown short isoform as a result of alternative promoter usage. Exon 2 of CPVL is drastically reduced in NCTX, while exons 5–6 are expressed in a complementary pattern (C). A more than 10-fold enrichment in STR of SKAP2 exon 13 is consistent with expression of a novel variant composed of exons 8–3 (D). (E) ROBO1 encodes two known isoforms, the full-length ROBO1a and the alternative short isoform ROBO1b. In the developing human NCTX, ROBO1a is highly enriched in temporal lobe, while ROBO1b is slightly enriched in PFC. (F) ANKRD32 is a previously uncharacterized gene that appears to encode two splice isoforms, ANKRD32a, which is evenly expressed across the NCTX, and ANKRD32b, which is significantly enriched in PFC. Bar graphs are mean±SEM. P-values represent the interaction between brain region or NCTX area and splice isoform.
Figure 7. Network Structure of Gene Co-Regulatory Relationships in Developing Human Neocortex
Network analysis was performed to identify modules of co-regulated genes. (A) Dendrogram showing clustering of genes based on topological overlap to identify modules of co-regulated genes in the NCTX. Modules were determined by dynamic tree cutting, numbered, and color-coded. (B–C) Heatmap (B) and first principal component (C) of expression data for genes in module M15 (cyan) suggest identification of the module with PFC. (D) The network structure of the PFC module illustrates which genes are the most interconnected. The PFC hub gene LOC400120 is an uncharacterized locus that was not DEX or DAS by conventional expression analysis. (E–G) Module M24 (orange), in contrast, corresponds to non-PFC areas. Hubs in this network include known forebrain patterning genes MEIS2 and FGFR1, as well as genes previously unknown or uncharacterized in nervous tissue, such as TRAM2 and C6orf65.
Comment in
- Transcriptional regulation and alternative splicing make for better brains.
Dehay C, Kennedy H. Dehay C, et al. Neuron. 2009 May 28;62(4):455-7. doi: 10.1016/j.neuron.2009.05.006. Neuron. 2009. PMID: 19477147
Similar articles
- Human-specific transcriptional networks in the brain.
Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM, Geschwind DH. Konopka G, et al. Neuron. 2012 Aug 23;75(4):601-17. doi: 10.1016/j.neuron.2012.05.034. Neuron. 2012. PMID: 22920253 Free PMC article. - Transcriptome Analysis Identifies Multifaceted Regulatory Mechanisms Dictating a Genetic Switch from Neuronal Network Establishment to Maintenance During Postnatal Prefrontal Cortex Development.
Kroeze Y, Oti M, van Beusekom E, Cooijmans RHM, van Bokhoven H, Kolk SM, Homberg JR, Zhou H. Kroeze Y, et al. Cereb Cortex. 2018 Mar 1;28(3):833-851. doi: 10.1093/cercor/bhw407. Cereb Cortex. 2018. PMID: 28108491 - Alternative splicing is frequent during early embryonic development in mouse.
Revil T, Gaffney D, Dias C, Majewski J, Jerome-Majewska LA. Revil T, et al. BMC Genomics. 2010 Jun 23;11:399. doi: 10.1186/1471-2164-11-399. BMC Genomics. 2010. PMID: 20573213 Free PMC article. - Deconstructing language by comparative gene expression: from neurobiology to microarray.
Oldham MC, Geschwind DH. Oldham MC, et al. Genes Brain Behav. 2006;5 Suppl 1:54-63. doi: 10.1111/j.1601-183X.2006.00195.x. Genes Brain Behav. 2006. PMID: 16417618 Review. - Alternative splicing: new insights from global analyses.
Blencowe BJ. Blencowe BJ. Cell. 2006 Jul 14;126(1):37-47. doi: 10.1016/j.cell.2006.06.023. Cell. 2006. PMID: 16839875 Review.
Cited by
- Cell-type-specific splicing of transcription regulators and Ptbp1 by Rbfox1/2/3 in the developing neocortex.
Ruan X, Hu K, Yang Y, Yang R, Tseng E, Kang B, Kauffman A, Zhong R, Zhang X. Ruan X, et al. bioRxiv [Preprint]. 2024 Sep 10:2024.09.09.612108. doi: 10.1101/2024.09.09.612108. bioRxiv. 2024. PMID: 39314274 Free PMC article. Updated. Preprint. - A polarized FGF8 source specifies frontotemporal signatures in spatially oriented cell populations of cortical assembloids.
Bosone C, Castaldi D, Burkard TR, Guzman SJ, Wyatt T, Cheroni C, Caporale N, Bajaj S, Bagley JA, Li C, Sorre B, Villa CE, Testa G, Krenn V, Knoblich JA. Bosone C, et al. Nat Methods. 2024 Nov;21(11):2147-2159. doi: 10.1038/s41592-024-02412-5. Epub 2024 Sep 18. Nat Methods. 2024. PMID: 39294368 Free PMC article. - Functional and regulatory diversification of Period genes responsible for circadian rhythm in vertebrates.
Kwak JS, León-Tapia MÁ, Diblasi C, Manousi D, Grønvold L, Sandvik GK, Saitou M. Kwak JS, et al. G3 (Bethesda). 2024 Oct 7;14(10):jkae162. doi: 10.1093/g3journal/jkae162. G3 (Bethesda). 2024. PMID: 39028850 Free PMC article. - Splicing-specific transcriptome-wide association uncovers genetic mechanisms for schizophrenia.
Hervoso JL, Amoah K, Dodson J, Choudhury M, Bhattacharya A, Quinones-Valdez G, Pasaniuc B, Xiao X. Hervoso JL, et al. Am J Hum Genet. 2024 Aug 8;111(8):1573-1587. doi: 10.1016/j.ajhg.2024.06.001. Epub 2024 Jun 25. Am J Hum Genet. 2024. PMID: 38925119 Free PMC article. - Autism patient-derived SHANK2BY29X mutation affects the development of ALDH1A1 negative dopamine neuron.
Lai W, Zhao Y, Chen Y, Dai Z, Chen R, Niu Y, Chen X, Chen S, Huang G, Shan Z, Zheng J, Hu Y, Chen Q, Gong S, Kang S, Guo H, Ma X, Song Y, Xia K, Wang J, Zhou L, So KF, Wang K, Qiu S, Zhang L, Chen J, Shi L. Lai W, et al. Mol Psychiatry. 2024 Oct;29(10):3180-3194. doi: 10.1038/s41380-024-02578-6. Epub 2024 May 4. Mol Psychiatry. 2024. PMID: 38704506 Free PMC article.
References
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. - PubMed
- Aruga J. The role of Zic genes in neural development. Mol. Cell Neurosci. 2004;26:205–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS054273/NS/NINDS NIH HHS/United States
- U24 NS051869/NS/NINDS NIH HHS/United States
- R01 NS054273-04/NS/NINDS NIH HHS/United States
- R01 MH060233-05/MH/NIMH NIH HHS/United States
- R37 MH060233/MH/NIMH NIH HHS/United States
- R01 NS056276/NS/NINDS NIH HHS/United States
- R56 MH060233/MH/NIMH NIH HHS/United States
- U24 NS051869-05/NS/NINDS NIH HHS/United States
- NS051869/NS/NINDS NIH HHS/United States
- R01 NS054273/NS/NINDS NIH HHS/United States
- U01 MH081896/MH/NIMH NIH HHS/United States
- R01 MH060233/MH/NIMH NIH HHS/United States
- MH060233/MH/NIMH NIH HHS/United States
- R01 NS056276-03/NS/NINDS NIH HHS/United States
- NS056276/NS/NINDS NIH HHS/United States
- MH081896/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources