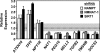Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters - PubMed (original) (raw)
Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters
Tong Zhang et al. J Biol Chem. 2009.
Abstract
In mammals, nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenylyltransferase 1 (NMNAT-1) constitute a nuclear NAD(+) salvage pathway which regulates the functions of NAD(+)-dependent enzymes such as the protein deacetylase SIRT1. One of the major functions of SIRT1 is to regulate target gene transcription through modification of chromatin-associated proteins. However, little is known about the molecular mechanisms by which NAD(+) biosynthetic enzymes regulate SIRT1 activity to control gene transcription in the nucleus. In this study we show that stable short hairpin RNA-mediated knockdown of NAMPT or NMNAT-1 in MCF-7 breast cancer cells reduces total cellular NAD(+) levels and alters global patterns of gene expression. Furthermore, we show that SIRT1 plays a key role in mediating the gene regulatory effects of NAMPT and NMNAT-1. Specifically, we found that SIRT1 binds to the promoters of genes commonly regulated by NAMPT, NMNAT-1, and SIRT1 and that SIRT1 histone deacetylase activity is regulated by NAMPT and NMNAT-1 at these promoters. Most significantly, NMNAT-1 interacts with, and is recruited to target gene promoters by SIRT1. Collectively, our results reveal a mechanism for the direct control of SIRT1 deacetylase activity at a set of target gene promoters by NMNAT-1. This mechanism, in collaboration with NAMPT-dependent regulation of nuclear NAD(+) production, establishes an important pathway for transcription regulation by NAD(+).
Figures
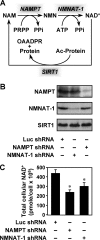
FIGURE 1.
Enzymes in the nuclear NAD+ salvage pathway regulate cellular NAD+ levels in MCF-7 cells. A, the nuclear NAD+ salvage pathway produces NAD+ for protein deacetylation by SIRT1. PRPP, phosphoribosylpyrophosphate; OAADPR, _O_-acetyl-ADP-ribose. B, stable shRNA-mediated knockdown of NMNAT-1 and NAMPT in MCF-7 cells. The Luc shRNA sequence was used as a control. NMNAT-1 and NAMPT protein levels were determined by Western blot analysis. C, total cellular NAD+ levels in control and NAMPT or NMNAT-1 knockdown cells. The concentrations of NAD+ in whole cell extracts were measured using a quantitative HPLC/mass spectrometry method with 18O standards. Error bars, S.E.; n = 8 independent biological replicates. *, significantly different from NAD+ levels in Luc control cells, p < 0.02 (Student's t test).
FIGURE 2.
NAMPT and NMNAT-1 regulate gene expression in MCF-7 cells. A, Venn diagram showing number of genes significantly regulated by NAMPT and NMNAT-1 knockdown (p < 0.05, Student's _t_ test) without a -fold change cutoff, as determined by expression microarray analysis. _B_, number of genes significantly regulated by NAMPT or NMNAT-1 knockdown (_p_ < 0.05, Student's _t_ test) after applying a -fold change cutoff (log2-fold change <−0.5 or >0.5). Numbers in parentheses indicate the percentage of total expressed genes. Each gene set is divided into up- and down-regulated groups. Note that two common genes are differentially regulated by NAMPT and NMNAT-1. C, scatter plot showing correlation of the expression profiles of 182 genes commonly regulated by NAMPT- and NMNAT-1 (i.e. the overlap in panel A). Spearman's rank correlation coefficient (r) and p value are indicated. D, effect of NAMPT, NMNAT-1, or SIRT1 knockdown with or without exogenously added NAD+ (1 m
m
) on the expression of NAMPT- and NMNAT-1-regulated genes. The target genes are selected based on the expression microarray analyses. Gene expression levels were determined by RT-qPCR using β-actin as the reference gene. Data are presented as expression levels relative to that in the Luc control cells. Error bars, S.E.; n ≥ 3 independent biological replicates.
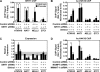
FIGURE 3.
Many NAMPT- and NMNAT-1-regulated genes show similar patterns of regulation by SIRT1 in MCF-7 cells. A, stable shRNA-mediated knockdown of SIRT1 in MCF-7 cells. SIRT1 protein levels were determined by Western blot analysis. The shRNA constructs that knockdown NAMPT and NMNAT-1 (Fig. 1_B_) have no effect on SIRT1 protein levels. B, number of genes significantly regulated by SIRT1 knockdown (p < 0.05, Student's t_ test) after applying a -fold change cutoff (log2-fold change <−0.5 or >0.5) as determined by expression microarray analysis. Numbers in parentheses indicate the percentage of total expressed genes. The gene set is divided into up- and down-regulated groups. C, expression profiles of the NAMPT- and NMNAT-1-regulated genes (Fig. 2_B) in response to SIRT1 knockdown. D, correlation analysis of the gene expression profiles shown in panel C. The plots show a 30-gene-moving average. Spearman's rank correlation coefficient (R) and p value are indicated.
FIGURE 4.
Regulation of target gene expression by NAMPT, NMNAT-1, and SIRT1 in MCF-7 cells. RT-qPCR confirmation of expression microarray data. Data are presented as expression levels relative to that in the luciferase control cells. The shaded area indicates the boundary for -fold change <1.414 (_i.e._ log2-fold change < 0.5) or -fold change >0.707 (i.e. log2-fold change >−0.5). Error bars, S.E.; n ≥ 3 independent biological replicates.
FIGURE 5.
NAMPT and NMNAT-1 regulate SIRT1 deacetylase activity at target gene promoters in MCF-7 cells. A, ChIP-qPCR analysis of SIRT1 localization at promoter and upstream (∼−10 kilobase) regions of target genes in MCF-7 cells. SIRT1 localization was also examined at the target gene promoters in SIRT1 knockdown cells. Statistical significance was determined by a two-tailed Student's t test (*, p < 0.05; **, p < 0.01). B and D, ChIP-qPCR analysis of acetylated H4K16 (Ac-H4K16) levels at target gene promoters in SIRT1, NAMPT, or NMNAT-1 knockdown cells or in control cells with or without sirtinol (50 μ
m
) treatment. Ac-H4K16 levels were normalized for H3 occupancy. Statistical significance for differences between Luc control samples and other samples was determined by a two-tailed Student's t test (*, p < 0.05; **, _p_ < 0.01). _C_, effect of sirtinol on the expression of SIRT1-regulated genes. RT-qPCR data are presented as expression levels relative to those in the control cells. The _shaded areas_ indicate the boundaries for -fold change <1.414 (_i.e._ log2-fold change < 0.5) or -fold change >0.707 (i.e. log2-fold change >−0.5). Statistical significance for differences between Luc control samples and other samples was determined by two-tailed Student's t test (*, p < 0.05; **, p < 0.01). All panels: error bars, S.E.; n ≥ 3 independent biological replicates.
FIGURE 6.
NMNAT-1 is recruited to target gene promoters by SIRT1. A, FLAG-based ChIP-qPCR analysis of NMNAT-1 localization at promoter and upstream (∼−10 kilobase) regions of target genes. MCF-7 cells expressing FLAG-NMNAT-1 were used for the FLAG ChIP assay. Error bars, S.E.; n ≥ 3 independent biological replicates. Statistical significance for differences between ChIP signals at upstream and promoter regions of the same gene was determined by a two-tailed Student's t test (*, p < 0.05; **, p < 0.01). B, FLAG-based ChIP-qPCR analysis of NMNAT-1 localization at target gene promoters in control or SIRT1 knockdown cells. Error bars, S.E.; n ≥ 3 independent biological replicates. Statistical significance for differences between ChIP signals in control and SIRT1 knockdown cells was determined by a two-tailed Student's t test (*, p < 0.05; **, p < 0.01). C, ChIP-Western analysis of interaction between SIRT1 and NMNAT-1. MCF-7 cells expressing GFP or FLAG-NMNAT-1 were used for FLAG and SIRT1 ChIP assays. The immunoprecipitated (IP) material was subjected to SIRT1 and FLAG Western blot analysis, respectively. Gels shown are representative of three independent experiments. D, GST-NMNAT-1 interaction assay with native SIRT1 from nuclear extract. SIRT1 bound to glutathione-agarose resin was detected by Western blot analysis. The gel shown is representative of three independent experiments.
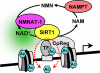
FIGURE 7.
A model for the regulation of SIRT1 activity at target gene promoters by NAMPT and NMNAT-1. SIRT1 binds to target gene promoters and regulates the acetylation status of transcription factors, histones, and other chromatin-associated proteins in an NAD+-dependent manner. Both NAMPT and NMNAT-1 localize to the nucleus and constitute an NAD+ recycling pathway. Nuclear NAD+ production by NAMPT and NMNAT-1 supports SIRT1 deacetylase activity. In addition, NMNAT-1 interacts with SIRT1 and is recruited to SIRT1 target gene promoters. Interactions between NMNAT-1 and SIRT1 on chromatin may underlie novel mechanisms for transcriptional regulation by SIRT1. TF, transcription factor. CoReg, coregulator.
Similar articles
- SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions.
Zhang T, Kraus WL. Zhang T, et al. Biochim Biophys Acta. 2010 Aug;1804(8):1666-75. doi: 10.1016/j.bbapap.2009.10.022. Epub 2009 Oct 30. Biochim Biophys Acta. 2010. PMID: 19879981 Free PMC article. Review. - The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells.
Revollo JR, Grimm AA, Imai S. Revollo JR, et al. J Biol Chem. 2004 Dec 3;279(49):50754-63. doi: 10.1074/jbc.M408388200. Epub 2004 Sep 20. J Biol Chem. 2004. PMID: 15381699 - Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT.
Choi SE, Fu T, Seok S, Kim DH, Yu E, Lee KW, Kang Y, Li X, Kemper B, Kemper JK. Choi SE, et al. Aging Cell. 2013 Dec;12(6):1062-72. doi: 10.1111/acel.12135. Epub 2013 Aug 11. Aging Cell. 2013. PMID: 23834033 Free PMC article. - Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1.
Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Nakahata Y, et al. Science. 2009 May 1;324(5927):654-7. doi: 10.1126/science.1170803. Epub 2009 Mar 12. Science. 2009. PMID: 19286518 Free PMC article.
Cited by
- Sirtuin 1-mediated effects of exercise and resveratrol on mitochondrial biogenesis.
Menzies KJ, Singh K, Saleem A, Hood DA. Menzies KJ, et al. J Biol Chem. 2013 Mar 8;288(10):6968-79. doi: 10.1074/jbc.M112.431155. Epub 2013 Jan 17. J Biol Chem. 2013. PMID: 23329826 Free PMC article. - On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1.
Luo X, Kraus WL. Luo X, et al. Genes Dev. 2012 Mar 1;26(5):417-32. doi: 10.1101/gad.183509.111. Genes Dev. 2012. PMID: 22391446 Free PMC article. Review. - Histone deacetylase inhibitors: clinical implications for hematological malignancies.
Tambaro FP, Dell'aversana C, Carafa V, Nebbioso A, Radic B, Ferrara F, Altucci L. Tambaro FP, et al. Clin Epigenetics. 2010 Sep;1(1-2):25-44. doi: 10.1007/s13148-010-0006-2. Epub 2010 Jul 28. Clin Epigenetics. 2010. PMID: 22704087 Free PMC article. - SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions.
Zhang T, Kraus WL. Zhang T, et al. Biochim Biophys Acta. 2010 Aug;1804(8):1666-75. doi: 10.1016/j.bbapap.2009.10.022. Epub 2009 Oct 30. Biochim Biophys Acta. 2010. PMID: 19879981 Free PMC article. Review. - Regulation of SIRT1 in cellular functions: role of polyphenols.
Chung S, Yao H, Caito S, Hwang JW, Arunachalam G, Rahman I. Chung S, et al. Arch Biochem Biophys. 2010 Sep 1;501(1):79-90. doi: 10.1016/j.abb.2010.05.003. Epub 2010 May 5. Arch Biochem Biophys. 2010. PMID: 20450879 Free PMC article. Review.
References
- Berger F., Ramírez-Hernández M. H., Ziegler M. (2004) Trends Biochem. Sci. 29, 111–118 - PubMed
- Rongvaux A., Andris F., Van Gool F., Leo O. (2003) BioEssays 25, 683–690 - PubMed
- Rongvaux A., Shea R. J., Mulks M. H., Gigot D., Urbain J., Leo O., Andris F. (2002) Eur. J. Immunol. 32, 3225–3234 - PubMed
- Kitani T., Okuno S., Fujisawa H. (2003) FEBS Lett. 544, 74–78 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous