Localisation of DivIVA by targeting to negatively curved membranes - PubMed (original) (raw)
Localisation of DivIVA by targeting to negatively curved membranes
Rok Lenarcic et al. EMBO J. 2009.
Abstract
DivIVA is a conserved protein in Gram-positive bacteria and involved in various processes related to cell growth, cell division and spore formation. DivIVA is specifically targeted to cell division sites and cell poles. In Bacillus subtilis, DivIVA helps to localise other proteins, such as the conserved cell division inhibitor proteins, MinC/MinD, and the chromosome segregation protein, RacA. Little is known about the mechanism that localises DivIVA. Here we show that DivIVA binds to liposomes, and that the N terminus harbours the membrane targeting sequence. The purified protein can stimulate binding of RacA to membranes. In mutants with aberrant cell shapes, DivIVA accumulates where the cell membrane is most strongly curved. On the basis of electron microscopic studies and other data, we propose that this is due to molecular bridging of the curvature by DivIVA multimers. This model may explain why DivIVA localises at cell division sites. A Monte-Carlo simulation study showed that molecular bridging can be a general mechanism for binding of proteins to negatively curved membranes.
Figures
Figure 1
Amino-acid alignment of DivIVA homologues (A) from the following: B. subtilis (B. sub.), S. aureus (S. aur.), S. pneumoniae (S. pneu.), M. tuberculosis (M. tub.) and S. coelicolor (S. coe.). Homologues and similar amino acids are boxed. The length of the C termini that extend beyond the B. subtilis DivIVA sequence is indicated. Presented above the sequence is the secondary structure prediction for B. subtilis DivIVA: c, coiled; h, helix. (B) Amphipathic helix prediction from the LOCATE program. The amino-acid positions are indicated and the Y axis shows the hydrophobic moments of the putative amphipathic helices.
Figure 2
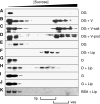
DivIVA-membrane interactions analysed with sucrose density gradient centrifugation. Gradient fractions were analysed by western blotting using GFP- or DivIVA-specific antibodies. The top fractions (low density) to the bottom fractions (high density) run from left to right. DivIVA-GFP (DG) was mixed with membrane vesicles (V; B) that were either incubated at high-salt concentrations (V-salt; C) or treated with Proteinase K (V-prot; D). Alternatively, purified DivIVA-GFP (DG), DivIVA (D) or GFP (G) was incubated with liposomes (Lip; F, H, and J). In all experiments, an excess of BSA (BSA) was present, which is indicated in the Coomassie staining of a blot (K). (A, E, G and I) The results of protein mixtures that contained no membrane vesicles or liposomes. Membranes vesicles (ves) and liposomes (lip) were clearly visible, and their position in the gradients is indicated below the figure.
Figure 3
Binding of purified GFP (A), DivIVA-GFP (B) and DivIVA (C) to liposomes adhered to a Biacore L1 sensor chip. Protein samples were injected (a), and after ∼2 min, followed by an injection of buffer alone (b). The chip was regenerated by a short injection (c and d) with 0.1 M NaOH solution. The flow rate was 30 μl/min, and protein concentrations were 3.1, 1.1 and 1.5 mg/ml for GFP, DivIVA-GFP and DivIVA, respectively. The response is given in artificial resonance units (RU). (D–G) SPR analysis of DivIVA deletions that were purified as MBP fusions. The C-terminal deletion (ΔC-DivIVA-MBP) lacks the last 20 amino acids of DivIVA and the N-terminal deletion (ΔN-DivIVA-MBP) lacks the first 40 amino acids of DivIVA. Protein concentrations were 0.4 mg/ml. (H) Sedimentation analyses of the N- and C-terminal DivIVA deletions in the presence and absence of liposomes. The total fraction, before centrifugation (T), and the supernatant (S) and pellet (P) fractions after centrifugation, was analysed by SDS–PAGE.
Figure 4
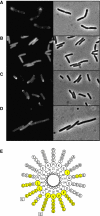
Fluorescence microscopy of cells expressing different DivIVA-GFP deletion constructs: (A) E. coli cells expressing full-length DivIVA fused to GFP, (B) E. coli cells expressing amino acids 1–40 of DivIVA fused to GFP, (C) E. coli cells expressing amino acids 1–60 of DivIVA fused to GFP and (D) B. subtilis cells expressing amino acids 1–60 of DivIVA fused to GFP (expression was induced by xylose). The latter strain (B. subtilis LH60) contains a deletion of the wild-type divIVA and minCD genes, which results in polar division and minicells. (E) Helical wheel projection of the amphipathic helix located at amino acids 22–41 of DivIVA. Hydrophobic residues are marked in yellow. The inner circle represents the amino-acid residues of DivIVA from B. subtilis. The amino-acid residues of S. aureus, S. pneumoniae, M. turberculosis and S. coelicolor DivIVA are depicted from the second (inner) to the fifth (outer) circle, respectively. Mutations in hydrophobic residues (V25E or L29E) are indicated.
Figure 5
Electron micrographs of negatively stained liposomes (A), and liposomes mixed with DivIVA (B, C). Liposomes were extruded using a 400-nm pore filter. Inset in panel A is a 2 × enlargement. Panel C is a 3 × enlargement of panel B, to show the layered structure due to stacking of liposomes. Scale bars=200 nm. Electron micrographs of thin sections of liposomes (D), and liposomes mixed with DivIVA (E–I). Liposomes were extruded using a 100-nm pore filter. Arrows indicate cruciform-like structures. (I) Part of a large liposome to which a cluster of DivIVA proteins (P) is bound; arrows point to DivIVA stalks attached to the membrane (M). Scale bars=200 nm in panels D and E, and 100 nm in panels F–I.
Figure 6
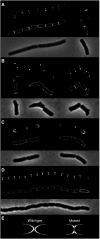
Localisation of DivIVA-GFP in cells of different shape: (A) wild-type B. subtilis cells, (B) compilation of several deformed B. subtilis mreB mutant cells (B. subtilis 3292), (C) wild-type E. coli cells and (D) E. coli murein hydrolase mutant cells (E. coli MHD63). To increase the resolution, deconvoluted images of Z-stacks are shown. The upper panels show GFP fluorescence, the middle panels show fluorescent membrane stain and the lower panels show phase-contrast images. (E) A schematic representation of the poles of wild-type E. coli cells and the murein hydrolase mutant (grey). DivIVA-GFP is indicated in white.
Figure 7
Bacterial two-hybrid interaction assay (A). divIVA and racA were cloned in different expression vectors, and the combinations screened for adenylate cyclase activity (blue colonies). A positive interaction was observed with a DivIVA-adenylate cyclase T25 fragment on the low-copy plasmid p25-N and an adenylate cyclase T18 fragment-RacA fusion on the high-copy plasmid pUT18C. pKT25: low-copy plasmid for N-terminal adenylate cyclase T25 fragment fusion, p25-N: low-copy plasmid for C-terminal adenylate cyclase fusion, pUT18C: high-copy plasmid for N-terminal adenylate cyclase T18 fragment fusion, pUT18: high-copy plasmid for C-terminal adenylate cyclase T18 fragment fusion. The effect of DivIVA on the binding of MBP-RacA to liposomes (B). MBP-RacA (6.4 ng) and DivIVA (3.6 μg) were mixed with liposomes (90 μg) in a sucrose-containing buffer, and loaded at the bottom of a sucrose gradient. After centrifugation, gradients where sampled in five fractions (top fractions (low density) to bottom fractions (high density) run from left to right). Liposomes floated to the two top fractions and where clearly visible. Gradient fractions were analysed by western blotting using RacA-specific antibodies.
Figure 8
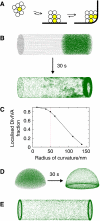
Schematic presentation of the molecular bridging model for the formation of stable DivIVA clusters at negatively curved cell membranes (A). DivIVA oligomers are indicated as spheres that form a free-floating cluster (left), a cluster that attaches to the cell membrane (middle) and a cluster that fills the corner of a curved membrane (right). Oligomers that can detach and diffuse away are transparent, and oligomers that are enclosed are indicated in yellow (see main text for more details). (B) Monte-Carlo simulation of 200 spheres that diffuse freely in a cylindrical volume, representing a rod-like bacterium. The pictures show the distribution over 25k iterations. The spheres depicting DivIVA oligomers (green) have a radius of 12.5 nm, and the dimensions of the cylinder are 4 × 1 μm (length × diameter). The curvature of the membranes at the transition from lateral wall of the cylinder to the sides has a radius of 50 nm. In this simulation, the spheres can make eight contacts, with two membrane contacts maximal (by weighing a membrane interaction as four contacts). _E_pp and _E_pm were 3.5 kBT and 5.5 kBT. To reduce cpu time, we started with an asymmetric distribution (the simulation took 5 days on a dual-core Intel processor). (C) Localisation of 12.5 nm spheres in relation to the curvature of the membrane. The simulation conditions were the same as in panel B. To quantify the localisation, we define a cutoff distance, typically 150 nm, and assume that a sphere is localised at the high curvature region when it is within this cutoff distance. (D) Monte-Carlo simulation of 200 spheres diffusing in a hemispheric volume with a diameter of 1 μm, representing a dividing coccoid. The same constants were used as in panel B except that in this case the spheres can make only four contacts, and there are no restrains on the number of membrane contacts. (E) Monte-Carlo simulation of rod-shaped structures. The simulation conditions were the same as in panel B except that only 100 rods were used to limit cpu time.
Comment in
- Irresistible curves.
Juarez JR, Margolin W. Juarez JR, et al. EMBO J. 2009 Aug 5;28(15):2147-8. doi: 10.1038/emboj.2009.167. EMBO J. 2009. PMID: 19654604 Free PMC article. No abstract available.
Similar articles
- Protein-protein interaction domains of Bacillus subtilis DivIVA.
van Baarle S, Celik IN, Kaval KG, Bramkamp M, Hamoen LW, Halbedel S. van Baarle S, et al. J Bacteriol. 2013 Mar;195(5):1012-21. doi: 10.1128/JB.02171-12. Epub 2012 Dec 21. J Bacteriol. 2013. PMID: 23264578 Free PMC article. - Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis.
Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, Pogliano J, Ramamurthi KS. Eswaramoorthy P, et al. mBio. 2011 Nov 22;2(6):e00257-11. doi: 10.1128/mBio.00257-11. Print 2011. mBio. 2011. PMID: 22108385 Free PMC article. - Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast.
Edwards DH, Thomaides HB, Errington J. Edwards DH, et al. EMBO J. 2000 Jun 1;19(11):2719-27. doi: 10.1093/emboj/19.11.2719. EMBO J. 2000. PMID: 10835369 Free PMC article. - Finding the corners in a cell.
Strahl H, Hamoen LW. Strahl H, et al. Curr Opin Microbiol. 2012 Dec;15(6):731-6. doi: 10.1016/j.mib.2012.10.006. Epub 2012 Nov 22. Curr Opin Microbiol. 2012. PMID: 23182676 Review. - Structural basis for interaction of DivIVA/GpsB proteins with their ligands.
Halbedel S, Lewis RJ. Halbedel S, et al. Mol Microbiol. 2019 Jun;111(6):1404-1415. doi: 10.1111/mmi.14244. Epub 2019 Apr 2. Mol Microbiol. 2019. PMID: 30887576 Review.
Cited by
- The positioning of the asymmetric septum during sporulation in Bacillus subtilis.
Barák I, Muchová K. Barák I, et al. PLoS One. 2018 Aug 9;13(8):e0201979. doi: 10.1371/journal.pone.0201979. eCollection 2018. PLoS One. 2018. PMID: 30092000 Free PMC article. - How do bacteria localize proteins to the cell pole?
Laloux G, Jacobs-Wagner C. Laloux G, et al. J Cell Sci. 2014 Jan 1;127(Pt 1):11-9. doi: 10.1242/jcs.138628. Epub 2013 Dec 17. J Cell Sci. 2014. PMID: 24345373 Free PMC article. Review. - Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis.
Briley K Jr, Prepiak P, Dias MJ, Hahn J, Dubnau D. Briley K Jr, et al. Mol Microbiol. 2011 Jul;81(1):23-39. doi: 10.1111/j.1365-2958.2011.07695.x. Epub 2011 Jun 7. Mol Microbiol. 2011. PMID: 21564336 Free PMC article. - Identification of membrane curvature sensing motifs essential for VPS37A phagophore recruitment and autophagosome closure.
Ye Y, Liang X, Wang G, Bewley MC, Hamamoto K, Liu X, Flanagan JM, Wang HG, Takahashi Y, Tian F. Ye Y, et al. Commun Biol. 2024 Mar 15;7(1):334. doi: 10.1038/s42003-024-06026-7. Commun Biol. 2024. PMID: 38491121 Free PMC article. - DivIVA-mediated polar localization of ComN, a posttranscriptional regulator of Bacillus subtilis.
dos Santos VT, Bisson-Filho AW, Gueiros-Filho FJ. dos Santos VT, et al. J Bacteriol. 2012 Jul;194(14):3661-9. doi: 10.1128/JB.05879-11. Epub 2012 May 11. J Bacteriol. 2012. PMID: 22582279 Free PMC article.
References
- Allen MP, Tildesley DJ (1989) Computer Simulation of Liquids. Oxford: Oxford University Press
- Anderluh G, Besenicar M, Kladnik A, Lakey JH, Macek P (2005) Properties of nonfused liposomes immobilized on an L1 Biacore chip and their permeabilization by a eukaryotic pore-forming toxin. Anal Biochem 344: 43–52 - PubMed
- Ben-Yehuda S, Rudner DZ, Losick R (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 - PubMed
- Binder K, Heermann D (2002) Monte Carlo Simulation in Statistical Physics: An Introduction, 4th edn. Berlin: Springer-Verlag.
- Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J (2008) A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol 70: 1556–1569 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases