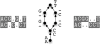Fast statistical alignment - PubMed (original) (raw)
Fast statistical alignment
Robert K Bradley et al. PLoS Comput Biol. 2009 May.
Abstract
We describe a new program for the alignment of multiple biological sequences that is both statistically motivated and fast enough for problem sizes that arise in practice. Our Fast Statistical Alignment program is based on pair hidden Markov models which approximate an insertion/deletion process on a tree and uses a sequence annealing algorithm to combine the posterior probabilities estimated from these models into a multiple alignment. FSA uses its explicit statistical model to produce multiple alignments which are accompanied by estimates of the alignment accuracy and uncertainty for every column and character of the alignment--previously available only with alignment programs which use computationally-expensive Markov Chain Monte Carlo approaches--yet can align thousands of long sequences. Moreover, FSA utilizes an unsupervised query-specific learning procedure for parameter estimation which leads to improved accuracy on benchmark reference alignments in comparison to existing programs. The centroid alignment approach taken by FSA, in combination with its learning procedure, drastically reduces the amount of false-positive alignment on biological data in comparison to that given by other methods. The FSA program and a companion visualization tool for exploring uncertainty in alignments can be used via a web interface at http://orangutan.math.berkeley.edu/fsa/, and the source code is available at http://fsa.sourceforge.net/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
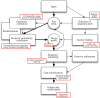
Figure 1. Overview of the components constituting the FSA alignment program.
The algorithms that are used in each component are highlighted in the accompanying boxes. The bold arrows show the simplest mode of use for FSA, where posterior probabilities are calculated directly using default parameters for all pairs of sequences and the optional steps of anchor finding and iterative refinement are omitted.
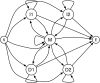
Figure 2. The default Pair HMM used by FSA.
By default FSA uses a Pair HMM with two sets of Insert (I) and Delete (D) states to generate a two-component geometric mixture distribution. FSA can optionally use a three-state HMM, which has only one set of Insert and Delete states. M is a Match state emitting aligned characters.
Figure 3. Two alignments (left and right) which make the same homology statements and therefore are both represented by the same POSET (center).
“The mathematics of distance-based alignment” in Text S1 discusses this view of alignments as POSETs. The alignment on the right minimizes the number of gap-open events and as such is appropriate for analyses such as inferring parsimonious indel frequencies across a clade. Alignments are displayed with TeXshade .
Figure 4. Schematic overview of FSA's parallelization strategy on a computer cluster.
For large input sizes, a disk-based database may be used to store some of the primary data structures and reduce memory usage.
Figure 5. The Java GUI allows users to visualize the estimated alignment accuracy under FSA's statistical model.
FSA's alignment is colored according the expected accuracy under FSA's statistical model (top) as well as according to the “true” accuracy (bottom) given from a comparison between FSA's alignment and the reference structural alignment. It is clear from inspection that accuracies estimated under FSA's statistical model correspond closely to the true accuracies. Sequences are from alignment BBS12030 in the RV12 dataset of BAliBASE 3 .
Similar articles
- Bayesian coestimation of phylogeny and sequence alignment.
Lunter G, Miklós I, Drummond A, Jensen JL, Hein J. Lunter G, et al. BMC Bioinformatics. 2005 Apr 1;6:83. doi: 10.1186/1471-2105-6-83. BMC Bioinformatics. 2005. PMID: 15804354 Free PMC article. - Statistical alignment based on fragment insertion and deletion models.
Metzler D. Metzler D. Bioinformatics. 2003 Mar 1;19(4):490-9. doi: 10.1093/bioinformatics/btg026. Bioinformatics. 2003. PMID: 12611804 - MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities.
Liu Y, Schmidt B, Maskell DL. Liu Y, et al. Bioinformatics. 2010 Aug 15;26(16):1958-64. doi: 10.1093/bioinformatics/btq338. Epub 2010 Jun 23. Bioinformatics. 2010. PMID: 20576627 - Hidden Markov Models, grammars, and biology: a tutorial.
Mukherjee S, Mitra S. Mukherjee S, et al. J Bioinform Comput Biol. 2005 Apr;3(2):491-526. doi: 10.1142/s0219720005001077. J Bioinform Comput Biol. 2005. PMID: 15852517 Review. - Developments in Algorithms for Sequence Alignment: A Review.
Chao J, Tang F, Xu L. Chao J, et al. Biomolecules. 2022 Apr 6;12(4):546. doi: 10.3390/biom12040546. Biomolecules. 2022. PMID: 35454135 Free PMC article. Review.
Cited by
- A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar.
Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE Jr, Pritchard JK, Mason CE, Gilad Y. Perry GH, et al. Genome Biol Evol. 2012;4(2):126-35. doi: 10.1093/gbe/evr132. Epub 2011 Dec 7. Genome Biol Evol. 2012. PMID: 22155688 Free PMC article. - Accurate reconstruction of insertion-deletion histories by statistical phylogenetics.
Westesson O, Lunter G, Paten B, Holmes I. Westesson O, et al. PLoS One. 2012;7(4):e34572. doi: 10.1371/journal.pone.0034572. Epub 2012 Apr 20. PLoS One. 2012. PMID: 22536326 Free PMC article. - Efficient representation of uncertainty in multiple sequence alignments using directed acyclic graphs.
Herman JL, Novák Á, Lyngsø R, Szabó A, Miklós I, Hein J. Herman JL, et al. BMC Bioinformatics. 2015 Apr 1;16:108. doi: 10.1186/s12859-015-0516-1. BMC Bioinformatics. 2015. PMID: 25888064 Free PMC article. - Genome and life-history evolution link bird diversification to the end-Cretaceous mass extinction.
Berv JS, Singhal S, Field DJ, Walker-Hale N, McHugh SW, Shipley JR, Miller ET, Kimball RT, Braun EL, Dornburg A, Parins-Fukuchi CT, Prum RO, Winger BM, Friedman M, Smith SA. Berv JS, et al. Sci Adv. 2024 Aug 2;10(31):eadp0114. doi: 10.1126/sciadv.adp0114. Epub 2024 Jul 31. Sci Adv. 2024. PMID: 39083615 Free PMC article. - Evolutionary History of the Marchantia polymorpha Complex.
Linde AM, Sawangproh W, Cronberg N, Szövényi P, Lagercrantz U. Linde AM, et al. Front Plant Sci. 2020 Jun 26;11:829. doi: 10.3389/fpls.2020.00829. eCollection 2020. Front Plant Sci. 2020. PMID: 32670318 Free PMC article.
References
- Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, et al. Clustal Wand Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. - PubMed
- Edgar R, Batzoglou S. Multiple sequence alignment. Curr Opin Struct Biol. 2006;16:368–373. - PubMed
- Wong K, Suchard M, Huelsenbeck J. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases